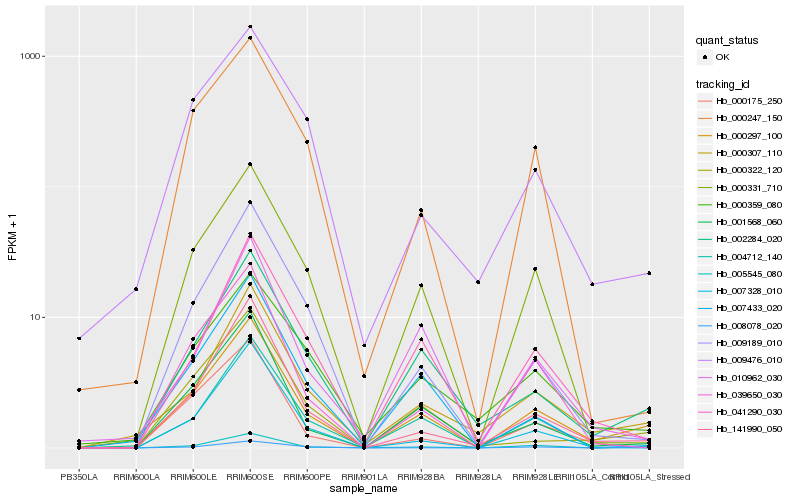
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000297_100 |
0.0 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 2 |
Hb_002284_020 |
0.1324709525 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 3 |
Hb_000307_110 |
0.1394567294 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 4 |
Hb_005545_080 |
0.1424021284 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |
| 5 |
Hb_001568_060 |
0.146270596 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 6 |
Hb_141990_050 |
0.1499275527 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 7 |
Hb_004712_140 |
0.1518247627 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 8 |
Hb_041290_030 |
0.1595989801 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 9 |
Hb_000331_710 |
0.1598500955 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000322_120 |
0.1609596083 |
- |
- |
PREDICTED: uncharacterized protein LOC105637900 [Jatropha curcas] |
| 11 |
Hb_008078_020 |
0.1685045413 |
- |
- |
hypothetical protein JCGZ_01297 [Jatropha curcas] |
| 12 |
Hb_039650_030 |
0.1724149656 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 13 |
Hb_009189_010 |
0.1735005258 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 14 |
Hb_000359_080 |
0.1774703547 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 15 |
Hb_010962_030 |
0.1793369035 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_007433_020 |
0.1795229625 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 17 |
Hb_000175_250 |
0.1828810281 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 18 |
Hb_000247_150 |
0.1835530072 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 19 |
Hb_007328_010 |
0.184943073 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 20 |
Hb_009476_010 |
0.185833916 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |