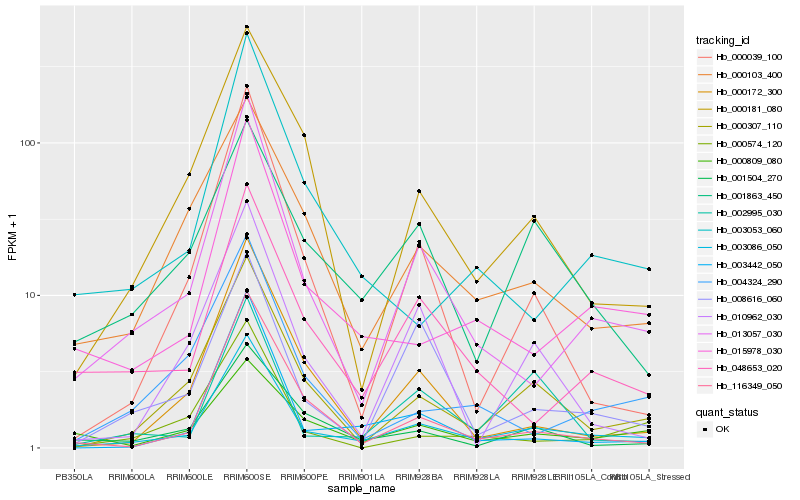
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003086_050 |
0.0 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000307_110 |
0.142615173 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 3 |
Hb_013057_030 |
0.1529024432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_015978_030 |
0.1559029701 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 5 |
Hb_116349_050 |
0.1748615021 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_048653_020 |
0.18002406 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000103_400 |
0.1816357462 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001504_270 |
0.185371626 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 9 |
Hb_003442_050 |
0.1907732063 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 10 |
Hb_000172_300 |
0.1924111719 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 11 |
Hb_000181_080 |
0.1926932966 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 12 |
Hb_001863_450 |
0.1936268943 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 13 |
Hb_000039_100 |
0.1959414521 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 14 |
Hb_000809_080 |
0.1977870778 |
- |
- |
50S ribosomal protein L25, putative [Ricinus communis] |
| 15 |
Hb_000574_120 |
0.1996318417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003053_060 |
0.2001954618 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 17 |
Hb_002995_030 |
0.2018442848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004324_290 |
0.2024312046 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 19 |
Hb_008616_060 |
0.2025830426 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 20 |
Hb_010962_030 |
0.2031295146 |
- |
- |
kinase, putative [Ricinus communis] |