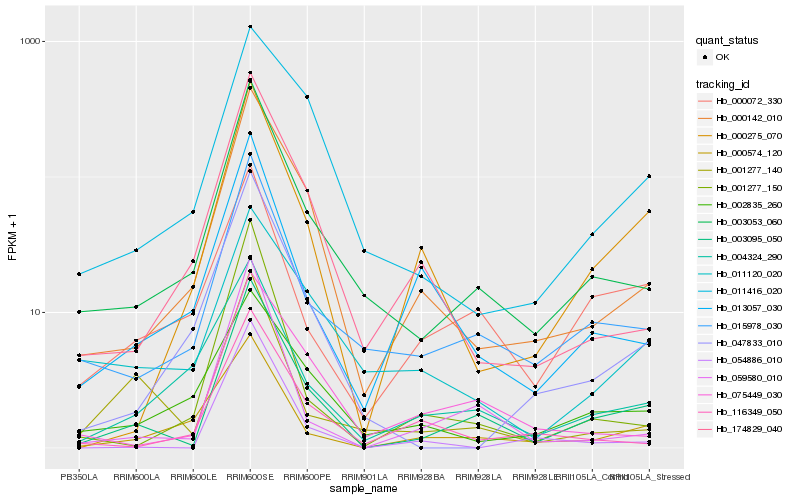
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003095_050 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 2 |
Hb_003053_060 |
0.1669155095 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 3 |
Hb_015978_030 |
0.1776687858 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 4 |
Hb_000142_010 |
0.1810934912 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 5 |
Hb_059580_010 |
0.1984808965 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 6 |
Hb_000275_070 |
0.2055302015 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 7 |
Hb_075449_030 |
0.2115890249 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 8 |
Hb_000072_330 |
0.2141243389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000574_120 |
0.2159815139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004324_290 |
0.2194892312 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 11 |
Hb_047833_010 |
0.2233741297 |
- |
- |
Avr9/Cf-9 rapidly elicited protein [Jatropha curcas] |
| 12 |
Hb_001277_150 |
0.2235544923 |
- |
- |
hypothetical protein VITISV_043492 [Vitis vinifera] |
| 13 |
Hb_011416_020 |
0.2242623135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_174829_040 |
0.2249796562 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 15 |
Hb_001277_140 |
0.2259512215 |
- |
- |
PREDICTED: plasma membrane calcium-transporting ATPase 2-like [Takifugu rubripes] |
| 16 |
Hb_013057_030 |
0.2275567479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_054886_010 |
0.228005177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_116349_050 |
0.2366332389 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_002835_260 |
0.2401812975 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 20 |
Hb_011120_020 |
0.2403774049 |
- |
- |
PREDICTED: casein kinase I-like isoform X1 [Jatropha curcas] |