| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
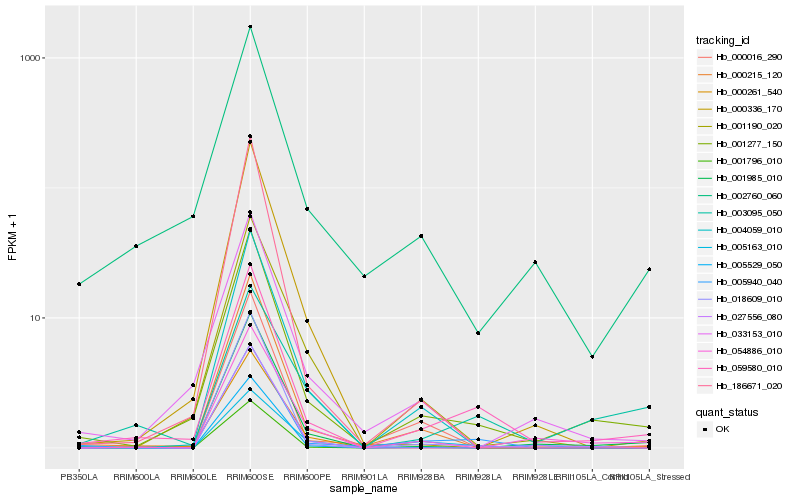
Hb_059580_010 |
0.0 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 2 |
Hb_001277_150 |
0.1092800079 |
- |
- |
hypothetical protein VITISV_043492 [Vitis vinifera] |
| 3 |
Hb_005940_040 |
0.1442205968 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 4 |
Hb_000215_120 |
0.1688263014 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 5 |
Hb_002760_060 |
0.1730976024 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 6 |
Hb_001985_010 |
0.1777139271 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_018609_010 |
0.1796413365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_033153_010 |
0.1826880299 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |
| 9 |
Hb_054886_010 |
0.1871096829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004059_010 |
0.1911814645 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 11 |
Hb_000336_170 |
0.1926653072 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 12 |
Hb_186671_020 |
0.1940160952 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_027556_080 |
0.1942150357 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g24130 [Jatropha curcas] |
| 14 |
Hb_005163_010 |
0.194894252 |
- |
- |
PREDICTED: probable terpene synthase 11 [Jatropha curcas] |
| 15 |
Hb_005529_050 |
0.1961819398 |
- |
- |
PREDICTED: cytochrome P450 94B3-like [Nelumbo nucifera] |
| 16 |
Hb_000016_290 |
0.1962224568 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 17 |
Hb_003095_050 |
0.1984808965 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 18 |
Hb_001796_010 |
0.1988751578 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 19 |
Hb_001190_020 |
0.1997945672 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000261_540 |
0.2008878289 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1-like [Jatropha curcas] |