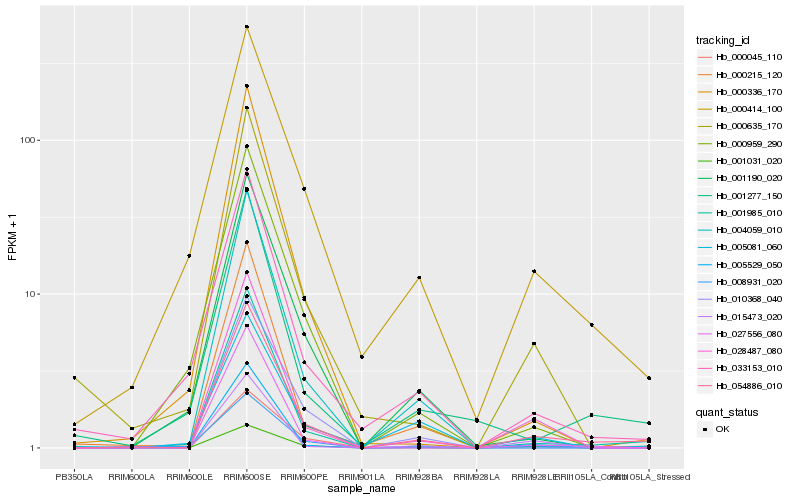
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054886_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001985_010 |
0.1039678975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_028487_080 |
0.1327728342 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000414_100 |
0.1435891324 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 5 |
Hb_027556_080 |
0.1447467438 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g24130 [Jatropha curcas] |
| 6 |
Hb_005529_050 |
0.1500569403 |
- |
- |
PREDICTED: cytochrome P450 94B3-like [Nelumbo nucifera] |
| 7 |
Hb_001277_150 |
0.1543151206 |
- |
- |
hypothetical protein VITISV_043492 [Vitis vinifera] |
| 8 |
Hb_000635_170 |
0.1551074016 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_001031_020 |
0.1554676714 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 10 |
Hb_000045_110 |
0.1563471523 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 11 |
Hb_004059_010 |
0.1585053685 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 12 |
Hb_000215_120 |
0.1591147484 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 13 |
Hb_001190_020 |
0.1596075616 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_010368_040 |
0.1609015564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_033153_010 |
0.1620521452 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |
| 16 |
Hb_015473_020 |
0.1623123078 |
- |
- |
hypothetical protein B456_001G090400 [Gossypium raimondii] |
| 17 |
Hb_005081_060 |
0.162661581 |
- |
- |
hypothetical protein EUGRSUZ_C00833 [Eucalyptus grandis] |
| 18 |
Hb_000336_170 |
0.1667964476 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 19 |
Hb_008931_020 |
0.1700941595 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 20 |
Hb_000959_290 |
0.1737272513 |
- |
- |
PREDICTED: uncharacterized protein LOC105636589 [Jatropha curcas] |