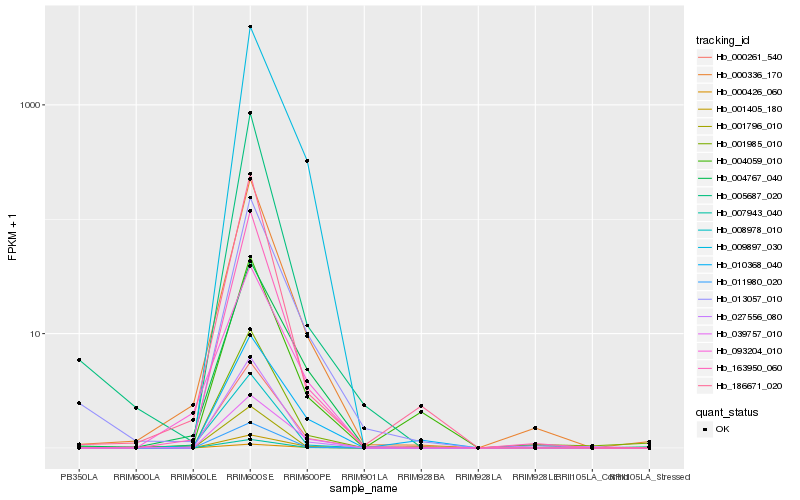
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007943_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_07271 [Jatropha curcas] |
| 2 |
Hb_009897_030 |
0.0416568185 |
- |
- |
- |
| 3 |
Hb_011980_020 |
0.0434591542 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_163950_060 |
0.0641449905 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_000336_170 |
0.0714720012 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 6 |
Hb_000261_540 |
0.0794734342 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1-like [Jatropha curcas] |
| 7 |
Hb_013057_010 |
0.0894552105 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 8 |
Hb_039757_010 |
0.0901525277 |
- |
- |
- |
| 9 |
Hb_001405_180 |
0.0940521216 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_027556_080 |
0.0966112244 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g24130 [Jatropha curcas] |
| 11 |
Hb_005687_020 |
0.1012860719 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 12 |
Hb_004767_040 |
0.1018256059 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 13 |
Hb_008978_010 |
0.1021790532 |
- |
- |
transmembrane receptor, putative [Ricinus communis] |
| 14 |
Hb_001796_010 |
0.1078103929 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 15 |
Hb_004059_010 |
0.1086400353 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 16 |
Hb_001985_010 |
0.1120482179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_186671_020 |
0.1156921983 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000426_060 |
0.1188167213 |
- |
- |
hypothetical protein CISIN_1g048192mg, partial [Citrus sinensis] |
| 19 |
Hb_010368_040 |
0.1208142208 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_093204_010 |
0.1227513749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |