| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
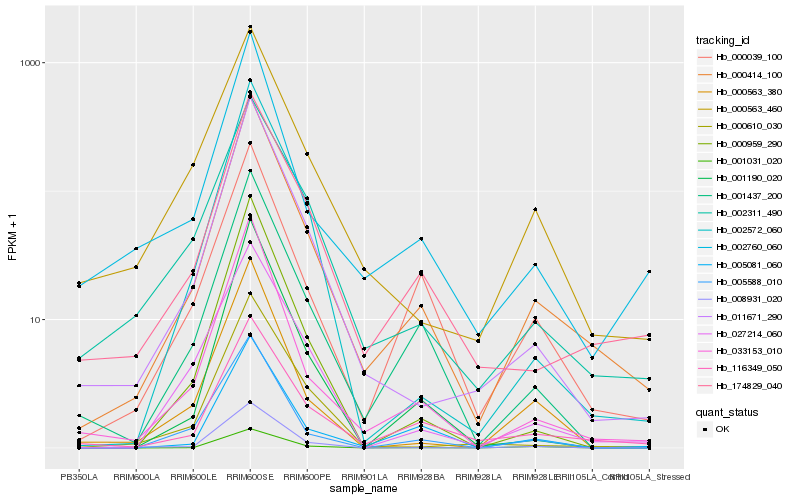
Hb_000414_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 2 |
Hb_033153_010 |
0.0964795916 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |
| 3 |
Hb_011671_290 |
0.1069475222 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 4 |
Hb_174829_040 |
0.1113589484 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 5 |
Hb_116349_050 |
0.1163523667 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_001437_200 |
0.1173197624 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 7 |
Hb_002572_060 |
0.128329812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008931_020 |
0.1296358428 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 9 |
Hb_000959_290 |
0.1310255839 |
- |
- |
PREDICTED: uncharacterized protein LOC105636589 [Jatropha curcas] |
| 10 |
Hb_002311_490 |
0.1317660552 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 11 |
Hb_000039_100 |
0.1326059957 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 12 |
Hb_001190_020 |
0.1330908466 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_005588_010 |
0.1340493088 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 14 |
Hb_002760_060 |
0.1350423971 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 15 |
Hb_000563_460 |
0.1355360059 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 16 |
Hb_027214_060 |
0.1360757451 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 17 |
Hb_001031_020 |
0.1362531394 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 18 |
Hb_000610_030 |
0.1396571891 |
- |
- |
hypothetical protein MIMGU_mgv1a0038491mg, partial [Erythranthe guttata] |
| 19 |
Hb_005081_060 |
0.1401523214 |
- |
- |
hypothetical protein EUGRSUZ_C00833 [Eucalyptus grandis] |
| 20 |
Hb_000563_380 |
0.1401739431 |
- |
- |
hypothetical protein CICLE_v10007206mg [Citrus clementina] |