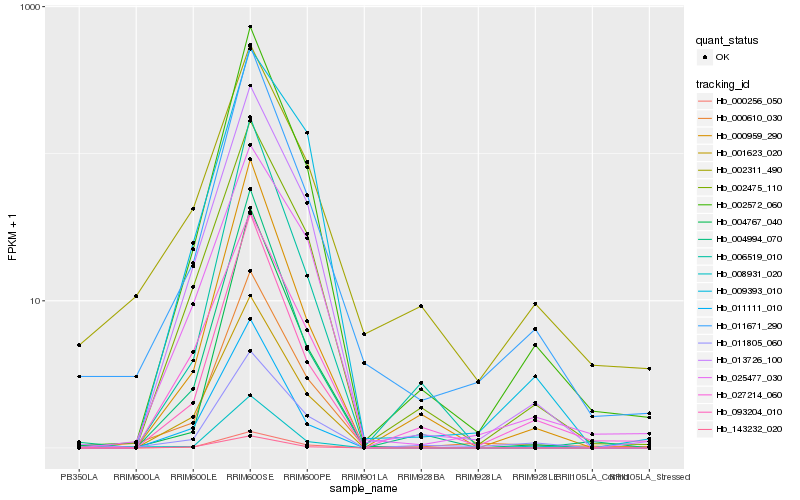
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000610_030 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a0038491mg, partial [Erythranthe guttata] |
| 2 |
Hb_011671_290 |
0.0688229142 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 3 |
Hb_002572_060 |
0.0711297653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_013726_100 |
0.0849750119 |
- |
- |
PREDICTED: BON1-associated protein 2 [Jatropha curcas] |
| 5 |
Hb_000256_050 |
0.0969904355 |
- |
- |
chitinase, putative [Ricinus communis] |
| 6 |
Hb_002475_110 |
0.1020139473 |
- |
- |
E3 ubiquitin ligase PUB14, putative [Ricinus communis] |
| 7 |
Hb_001623_020 |
0.1071729023 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 8 |
Hb_025477_030 |
0.1098905651 |
transcription factor |
TF Family: ERF |
dehydration-responsive element-binding protein 2 [Manihot esculenta] |
| 9 |
Hb_011805_060 |
0.1121347948 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_093204_010 |
0.1149898589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004767_040 |
0.1155971395 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_004994_070 |
0.1198020141 |
- |
- |
PREDICTED: BON1-associated protein 2 [Jatropha curcas] |
| 13 |
Hb_000959_290 |
0.1201306838 |
- |
- |
PREDICTED: uncharacterized protein LOC105636589 [Jatropha curcas] |
| 14 |
Hb_006519_010 |
0.1244764601 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 [Vitis vinifera] |
| 15 |
Hb_011111_010 |
0.1285870405 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21A [Jatropha curcas] |
| 16 |
Hb_027214_060 |
0.1290413078 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 17 |
Hb_143232_020 |
0.1299126395 |
- |
- |
hypothetical protein JCGZ_20467 [Jatropha curcas] |
| 18 |
Hb_009393_010 |
0.130304127 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 19 |
Hb_008931_020 |
0.1312312067 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 20 |
Hb_002311_490 |
0.1323267687 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |