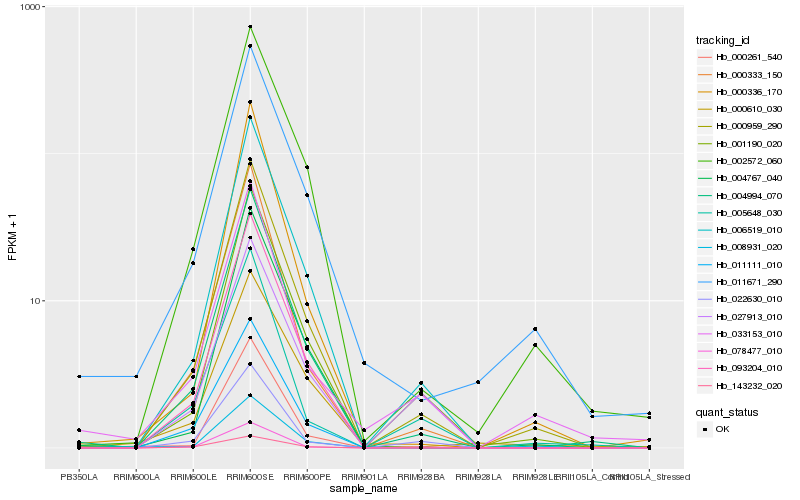
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000959_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636589 [Jatropha curcas] |
| 2 |
Hb_006519_010 |
0.0529408218 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 [Vitis vinifera] |
| 3 |
Hb_004994_070 |
0.0613563369 |
- |
- |
PREDICTED: BON1-associated protein 2 [Jatropha curcas] |
| 4 |
Hb_001190_020 |
0.0676525196 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_002572_060 |
0.070922714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_093204_010 |
0.0730562942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000333_150 |
0.0849753325 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 8 |
Hb_011111_010 |
0.0943590208 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21A [Jatropha curcas] |
| 9 |
Hb_004767_040 |
0.0981791785 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 10 |
Hb_000336_170 |
0.1044129292 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 11 |
Hb_011671_290 |
0.1066577242 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 12 |
Hb_078477_010 |
0.1075909748 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 13 |
Hb_000261_540 |
0.1135225725 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1-like [Jatropha curcas] |
| 14 |
Hb_033153_010 |
0.1155295668 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |
| 15 |
Hb_005648_030 |
0.1169365117 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH18-like [Jatropha curcas] |
| 16 |
Hb_008931_020 |
0.1174997112 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 17 |
Hb_143232_020 |
0.1185002633 |
- |
- |
hypothetical protein JCGZ_20467 [Jatropha curcas] |
| 18 |
Hb_027913_010 |
0.119525909 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_022630_010 |
0.1197251098 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000610_030 |
0.1201306838 |
- |
- |
hypothetical protein MIMGU_mgv1a0038491mg, partial [Erythranthe guttata] |