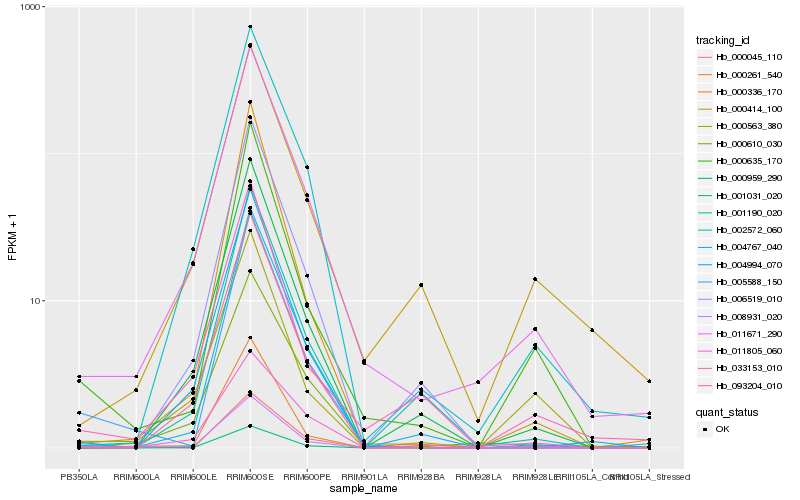
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008931_020 |
0.0 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 2 |
Hb_011671_290 |
0.0989087978 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 3 |
Hb_000563_380 |
0.1002378189 |
- |
- |
hypothetical protein CICLE_v10007206mg [Citrus clementina] |
| 4 |
Hb_000635_170 |
0.1002589121 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_000959_290 |
0.1174997112 |
- |
- |
PREDICTED: uncharacterized protein LOC105636589 [Jatropha curcas] |
| 6 |
Hb_000045_110 |
0.1181800689 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 7 |
Hb_002572_060 |
0.1200156918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001031_020 |
0.1243792582 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 9 |
Hb_000414_100 |
0.1296358428 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 10 |
Hb_000610_030 |
0.1312312067 |
- |
- |
hypothetical protein MIMGU_mgv1a0038491mg, partial [Erythranthe guttata] |
| 11 |
Hb_004767_040 |
0.1319718591 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_033153_010 |
0.1342098126 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |
| 13 |
Hb_011805_060 |
0.134378018 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 14 |
Hb_004994_070 |
0.1357777446 |
- |
- |
PREDICTED: BON1-associated protein 2 [Jatropha curcas] |
| 15 |
Hb_006519_010 |
0.1365900933 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 [Vitis vinifera] |
| 16 |
Hb_001190_020 |
0.1369099008 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000261_540 |
0.140108696 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1-like [Jatropha curcas] |
| 18 |
Hb_093204_010 |
0.1440818968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000336_170 |
0.144617615 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 20 |
Hb_005588_150 |
0.1492860448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |