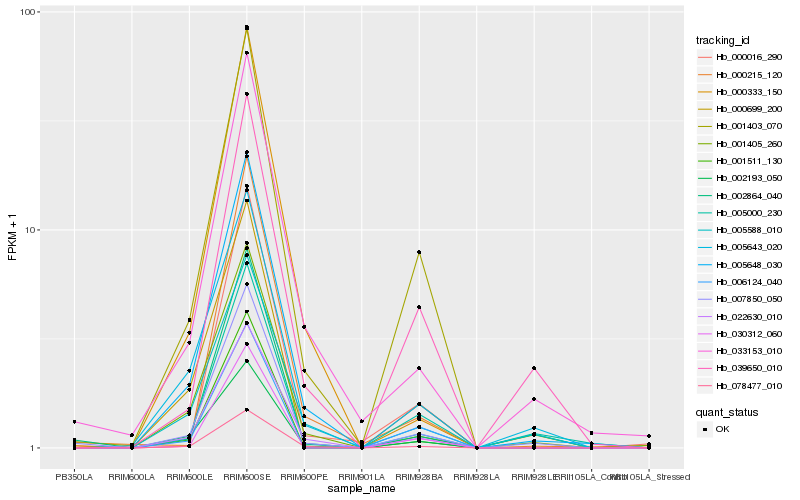
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007850_050 |
0.0 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_002864_040 |
0.0747392078 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 3 |
Hb_005648_030 |
0.1000957531 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH18-like [Jatropha curcas] |
| 4 |
Hb_001403_070 |
0.1120596283 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 5 |
Hb_033153_010 |
0.1150247862 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |
| 6 |
Hb_001405_260 |
0.1152131032 |
- |
- |
E3 ubiquitin ligase PUB14, putative [Ricinus communis] |
| 7 |
Hb_078477_010 |
0.116783479 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 8 |
Hb_022630_010 |
0.1220654762 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_005000_230 |
0.1264720488 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 10 |
Hb_001511_130 |
0.1268802798 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 11 |
Hb_005588_010 |
0.1317567402 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 12 |
Hb_005643_020 |
0.1345112881 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 13 |
Hb_039650_010 |
0.1376215335 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 14 |
Hb_002193_050 |
0.1385402103 |
- |
- |
PREDICTED: cytochrome P450 93A1-like [Citrus sinensis] |
| 15 |
Hb_000699_200 |
0.1409620173 |
- |
- |
hypothetical protein POPTR_0019s02340g [Populus trichocarpa] |
| 16 |
Hb_030312_060 |
0.1428272786 |
- |
- |
PREDICTED: uncharacterized protein LOC105634226 [Jatropha curcas] |
| 17 |
Hb_000215_120 |
0.1449425867 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 18 |
Hb_000333_150 |
0.1471825407 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 19 |
Hb_000016_290 |
0.1494173797 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 20 |
Hb_006124_040 |
0.1517198203 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |