| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
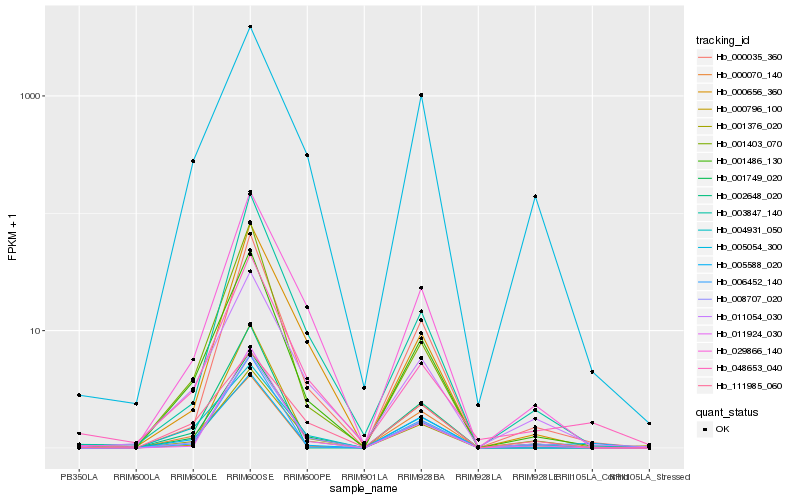
Hb_001486_130 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 2 |
Hb_001376_020 |
0.0680829381 |
- |
- |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 3 |
Hb_001749_020 |
0.0779990109 |
- |
- |
hypothetical protein CISIN_1g042220mg, partial [Citrus sinensis] |
| 4 |
Hb_008707_020 |
0.0930386336 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_001403_070 |
0.1025408768 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 6 |
Hb_004931_050 |
0.1064894388 |
transcription factor |
TF Family: MYB-related |
Transcription repressor MYB4, putative [Ricinus communis] |
| 7 |
Hb_000035_360 |
0.1137733476 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 8 |
Hb_029866_140 |
0.1142501027 |
- |
- |
PREDICTED: hevamine-A [Jatropha curcas] |
| 9 |
Hb_011054_030 |
0.1147130458 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 10 |
Hb_002648_020 |
0.1262211128 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Populus euphratica] |
| 11 |
Hb_005054_300 |
0.1277958269 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 12 |
Hb_005588_020 |
0.1331497198 |
- |
- |
PREDICTED: patatin-like protein 2 [Vitis vinifera] |
| 13 |
Hb_006452_140 |
0.1337238361 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 14 |
Hb_048653_040 |
0.1338951088 |
- |
- |
ankyrin repeat family protein [Populus trichocarpa] |
| 15 |
Hb_011924_030 |
0.1377729009 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_003847_140 |
0.1378841353 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000656_360 |
0.1410625112 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 18 |
Hb_000796_100 |
0.1439170854 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000070_140 |
0.1478675686 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 20 |
Hb_111985_060 |
0.1479060666 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |