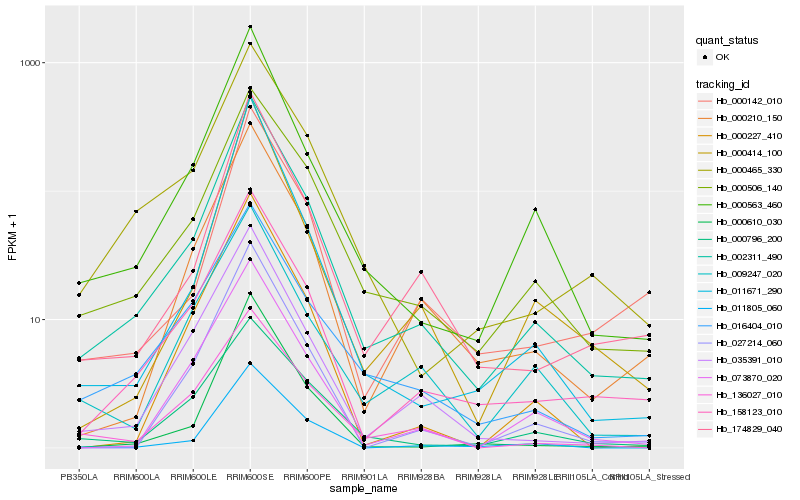
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_490 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 2 |
Hb_000506_140 |
0.0865974084 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 3 |
Hb_000563_460 |
0.0904604525 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 4 |
Hb_158123_010 |
0.094697679 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 5 |
Hb_174829_040 |
0.1041833489 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 6 |
Hb_000465_330 |
0.1073964605 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 7 |
Hb_035391_010 |
0.1081322804 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 8 |
Hb_000210_150 |
0.1132758334 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 9 |
Hb_011805_060 |
0.1194159702 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_016404_010 |
0.1196883883 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_027214_060 |
0.12144099 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 12 |
Hb_011671_290 |
0.1246628318 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 13 |
Hb_136027_010 |
0.1282266779 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 14 |
Hb_073870_020 |
0.1288923995 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000, partial [Jatropha curcas] |
| 15 |
Hb_000796_200 |
0.1293750642 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 3 [Jatropha curcas] |
| 16 |
Hb_000142_010 |
0.1317100904 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 17 |
Hb_000414_100 |
0.1317660552 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 18 |
Hb_009247_020 |
0.1321093919 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 19 |
Hb_000610_030 |
0.1323267687 |
- |
- |
hypothetical protein MIMGU_mgv1a0038491mg, partial [Erythranthe guttata] |
| 20 |
Hb_000227_410 |
0.132344712 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |