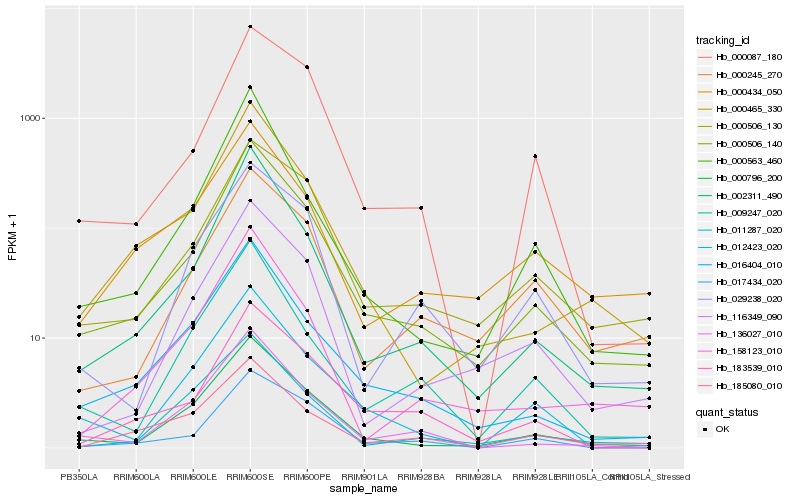
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000506_140 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 2 |
Hb_000796_200 |
0.0859032601 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 3 [Jatropha curcas] |
| 3 |
Hb_002311_490 |
0.0865974084 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 4 |
Hb_016404_010 |
0.109729407 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 5 |
Hb_000465_330 |
0.1115365644 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 6 |
Hb_000506_130 |
0.1149664996 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 7 |
Hb_116349_090 |
0.1213128788 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 8 |
Hb_158123_010 |
0.1243979787 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 9 |
Hb_183539_010 |
0.130202821 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 10 |
Hb_136027_010 |
0.1302757454 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_017434_020 |
0.130520328 |
transcription factor |
TF Family: C2H2 |
Zinc finger protein 4 [Populus trichocarpa] |
| 12 |
Hb_000563_460 |
0.1331835984 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 13 |
Hb_000087_180 |
0.137932352 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000245_270 |
0.1384143684 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 15 |
Hb_009247_020 |
0.1384506446 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 16 |
Hb_012423_020 |
0.1385967842 |
- |
- |
hypothetical protein JCGZ_20730 [Jatropha curcas] |
| 17 |
Hb_000434_050 |
0.1423897367 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 18 |
Hb_029238_020 |
0.1460323472 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 19 |
Hb_185080_010 |
0.1471373912 |
- |
- |
- |
| 20 |
Hb_011287_020 |
0.1476631183 |
- |
- |
ATP binding protein, putative [Ricinus communis] |