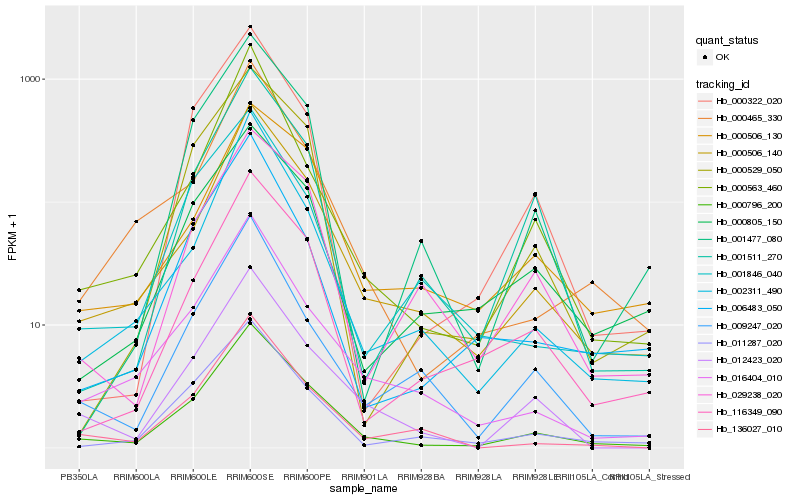
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000796_200 |
0.0 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 3 [Jatropha curcas] |
| 2 |
Hb_000506_140 |
0.0859032601 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 3 |
Hb_012423_020 |
0.1054652192 |
- |
- |
hypothetical protein JCGZ_20730 [Jatropha curcas] |
| 4 |
Hb_016404_010 |
0.1187904055 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 5 |
Hb_011287_020 |
0.121158003 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_116349_090 |
0.1244498294 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 7 |
Hb_136027_010 |
0.1280298538 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 8 |
Hb_002311_490 |
0.1293750642 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 9 |
Hb_000465_330 |
0.1310879176 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 10 |
Hb_000529_050 |
0.1356588725 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 11 |
Hb_009247_020 |
0.1364025891 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 12 |
Hb_000805_150 |
0.1394854385 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_001477_080 |
0.1404438573 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 14 |
Hb_006483_050 |
0.1415918898 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 15 |
Hb_000563_460 |
0.1420739212 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 16 |
Hb_000506_130 |
0.1421284754 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 17 |
Hb_029238_020 |
0.142311929 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 18 |
Hb_000322_020 |
0.1435472055 |
- |
- |
PREDICTED: uncharacterized protein LOC105637914 [Jatropha curcas] |
| 19 |
Hb_001511_270 |
0.1438142594 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 20 |
Hb_001846_040 |
0.1457224968 |
- |
- |
cytochrome P450, putative [Ricinus communis] |