| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
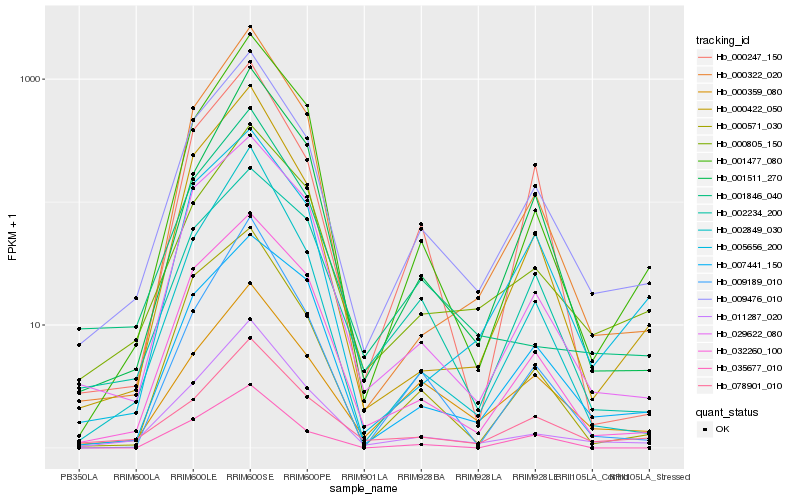
Hb_009476_010 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 2 |
Hb_011287_020 |
0.0819512778 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_032260_100 |
0.1030783107 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 4 |
Hb_000805_150 |
0.1032103569 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000422_050 |
0.1053236556 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 6 |
Hb_029622_080 |
0.107422438 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 7 |
Hb_078901_010 |
0.1110597126 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 8 |
Hb_000571_030 |
0.1118179095 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 9 |
Hb_005656_200 |
0.1196461965 |
- |
- |
Cation transport protein chaC, putative [Ricinus communis] |
| 10 |
Hb_001477_080 |
0.1204793392 |
- |
- |
hypothetical protein POPTR_0017s04860g [Populus trichocarpa] |
| 11 |
Hb_002849_030 |
0.1212410067 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 12 |
Hb_000247_150 |
0.1219700447 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 13 |
Hb_000322_020 |
0.1265553653 |
- |
- |
PREDICTED: uncharacterized protein LOC105637914 [Jatropha curcas] |
| 14 |
Hb_007441_150 |
0.1266330998 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 15 |
Hb_001511_270 |
0.1272168518 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 16 |
Hb_001846_040 |
0.128488056 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_009189_010 |
0.1307438309 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 18 |
Hb_000359_080 |
0.1309705718 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 19 |
Hb_002234_200 |
0.1321436535 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 20 |
Hb_035677_010 |
0.1353750043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |