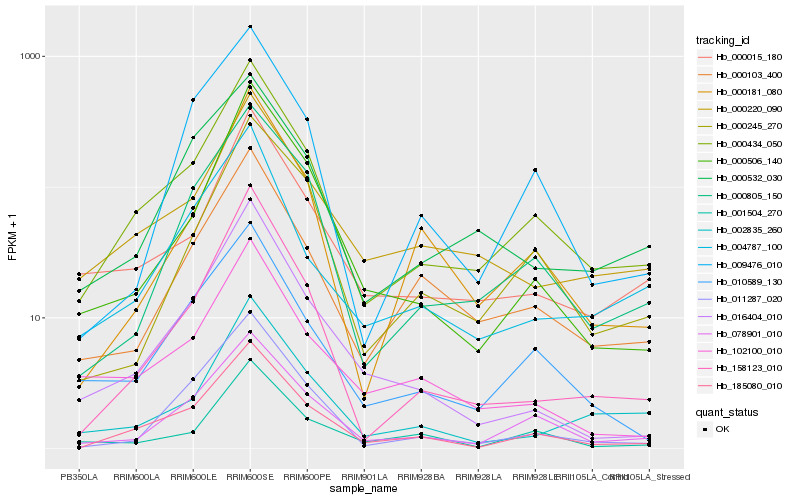
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000434_050 |
0.0 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 2 |
Hb_185080_010 |
0.09089984 |
- |
- |
- |
| 3 |
Hb_078901_010 |
0.1051315107 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 4 |
Hb_000015_180 |
0.1203817638 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 5 |
Hb_000805_150 |
0.1223749398 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_010589_130 |
0.1282597715 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_000220_090 |
0.1321631674 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 8 |
Hb_000103_400 |
0.1331577274 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_158123_010 |
0.1380807508 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 10 |
Hb_000532_030 |
0.1387957908 |
- |
- |
PREDICTED: oxalate--CoA ligase-like [Jatropha curcas] |
| 11 |
Hb_004787_100 |
0.1423543535 |
- |
- |
PREDICTED: transcription factor bHLH47 [Jatropha curcas] |
| 12 |
Hb_000506_140 |
0.1423897367 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 13 |
Hb_000245_270 |
0.1427630098 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 14 |
Hb_011287_020 |
0.1446932349 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000181_080 |
0.1448422936 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 16 |
Hb_002835_260 |
0.1483834162 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 17 |
Hb_102100_010 |
0.1551589334 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 18 |
Hb_001504_270 |
0.1563767119 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 19 |
Hb_009476_010 |
0.1567624583 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 20 |
Hb_016404_010 |
0.1594816734 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |