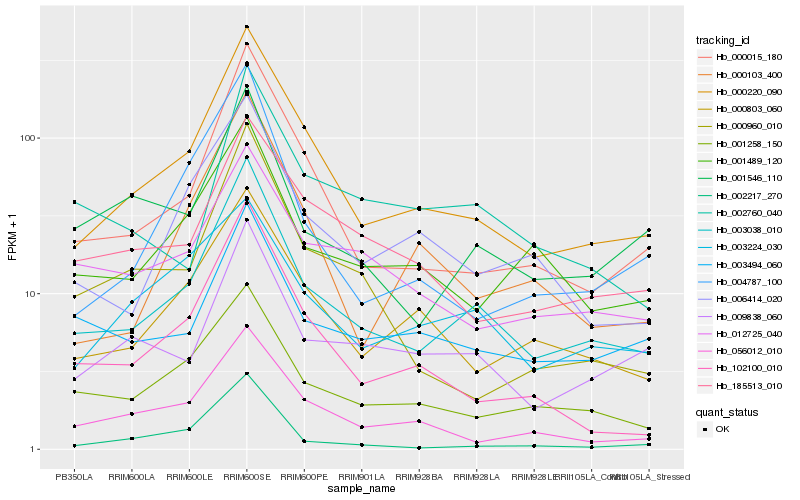
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003038_010 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_000220_090 |
0.1037068136 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 3 |
Hb_000015_180 |
0.1291123108 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 4 |
Hb_001258_150 |
0.1368519457 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 5 |
Hb_001546_110 |
0.1506376746 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 3 [Jatropha curcas] |
| 6 |
Hb_009838_060 |
0.1510028273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003494_060 |
0.1520500048 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000803_060 |
0.1551273248 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 9 |
Hb_006414_020 |
0.155955368 |
transcription factor |
TF Family: MYB |
- |
| 10 |
Hb_001489_120 |
0.1570583892 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_056012_010 |
0.1651521979 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 12 |
Hb_102100_010 |
0.1653680009 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 13 |
Hb_012725_040 |
0.1667210525 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_185513_010 |
0.1672537138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002760_040 |
0.1701668353 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 16 |
Hb_004787_100 |
0.1728164382 |
- |
- |
PREDICTED: transcription factor bHLH47 [Jatropha curcas] |
| 17 |
Hb_000103_400 |
0.176541386 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003224_030 |
0.1780528679 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 19 |
Hb_002217_270 |
0.1806703221 |
- |
- |
PREDICTED: desiccation-related protein PCC13-62-like [Jatropha curcas] |
| 20 |
Hb_000960_010 |
0.1819741402 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |