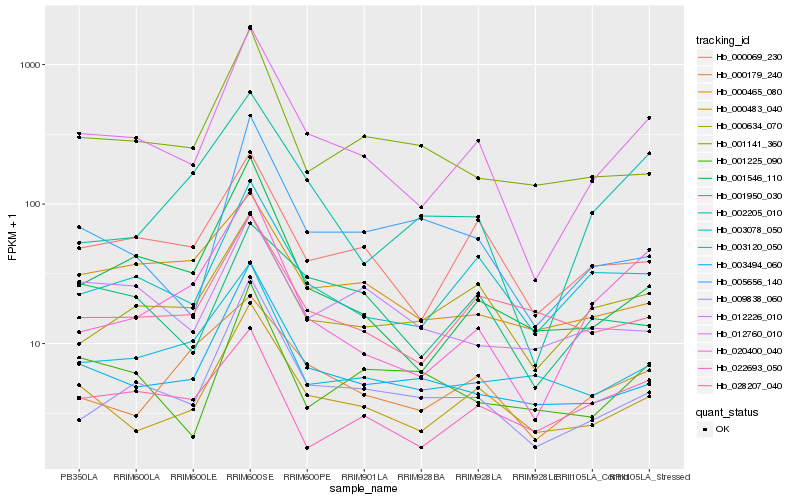
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012760_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000483_040 |
0.1249486515 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |
| 3 |
Hb_009838_060 |
0.1357921269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001546_110 |
0.1406115274 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 3 [Jatropha curcas] |
| 5 |
Hb_003078_050 |
0.1446932959 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 6 |
Hb_000069_230 |
0.1494013505 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 7 |
Hb_003494_060 |
0.151604151 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 8 |
Hb_020400_040 |
0.1668574136 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 9 |
Hb_000634_070 |
0.1686956366 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 10 |
Hb_005656_140 |
0.1770268267 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 11 |
Hb_028207_040 |
0.1793458118 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 12 |
Hb_001950_030 |
0.180526042 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 13 |
Hb_003120_050 |
0.1827613328 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 14 |
Hb_001141_360 |
0.1833781717 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 15 |
Hb_000465_080 |
0.1879774244 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 16 |
Hb_002205_010 |
0.1885325797 |
- |
- |
hypothetical protein JCGZ_03337 [Jatropha curcas] |
| 17 |
Hb_000179_240 |
0.1896016269 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001225_090 |
0.1941304975 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_012226_010 |
0.1954092113 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 20 |
Hb_022693_050 |
0.1969233231 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |