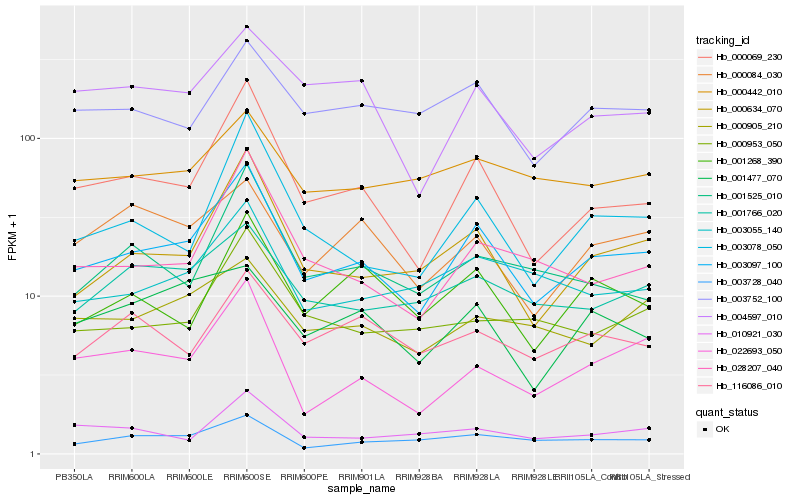
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003097_100 |
0.0 |
- |
- |
probable glycerol-3-phosphate dehydrogenase [NAD(+)] 1, cytosolic [Jatropha curcas] |
| 2 |
Hb_000069_230 |
0.0855002878 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 3 |
Hb_000634_070 |
0.101944546 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 4 |
Hb_003078_050 |
0.1173616001 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 5 |
Hb_028207_040 |
0.1259461625 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 6 |
Hb_022693_050 |
0.1278026548 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 7 |
Hb_003728_040 |
0.1321051385 |
- |
- |
PREDICTED: uncharacterized protein LOC101492305 [Cicer arietinum] |
| 8 |
Hb_003055_140 |
0.1325035971 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 9 |
Hb_004597_010 |
0.1325910278 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor MYC2 [Jatropha curcas] |
| 10 |
Hb_001766_020 |
0.1345952155 |
- |
- |
PREDICTED: probable galacturonosyltransferase 14 [Populus euphratica] |
| 11 |
Hb_003752_100 |
0.1360404595 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 12 |
Hb_001268_390 |
0.1366967211 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 13 |
Hb_000953_050 |
0.1408529645 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 14 |
Hb_001525_010 |
0.1425363908 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 15 |
Hb_000905_210 |
0.1430373519 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 16 |
Hb_010921_030 |
0.1448301306 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At2g01810 [Jatropha curcas] |
| 17 |
Hb_000084_030 |
0.1467108698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_116086_010 |
0.1470210711 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 19 |
Hb_000442_010 |
0.1471048667 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 20 |
Hb_001477_070 |
0.1485874138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |