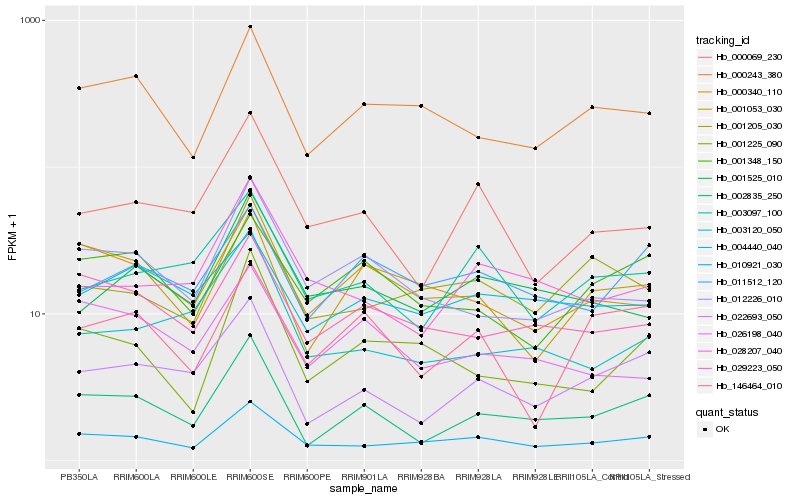
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002835_250 |
0.0 |
- |
- |
octicosapeptide/Phox/Bem1p domain-containing family protein [Populus trichocarpa] |
| 2 |
Hb_022693_050 |
0.1037828201 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 3 |
Hb_029223_050 |
0.1548309507 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010921_030 |
0.1594429289 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At2g01810 [Jatropha curcas] |
| 5 |
Hb_001053_030 |
0.1656190994 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000243_380 |
0.1667796725 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 7 |
Hb_012226_010 |
0.1688905851 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 8 |
Hb_000069_230 |
0.1744632156 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 9 |
Hb_001225_090 |
0.174764038 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_011512_120 |
0.1763202175 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 11 |
Hb_003120_050 |
0.1774683893 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 12 |
Hb_001348_150 |
0.1798824826 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 13 |
Hb_026198_040 |
0.1819283507 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 14 |
Hb_003097_100 |
0.1819803692 |
- |
- |
probable glycerol-3-phosphate dehydrogenase [NAD(+)] 1, cytosolic [Jatropha curcas] |
| 15 |
Hb_028207_040 |
0.1831465971 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 16 |
Hb_146464_010 |
0.184385738 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000340_110 |
0.1847578061 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 18 |
Hb_004440_040 |
0.1874137185 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_001205_030 |
0.1896514044 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0007s12970g [Populus trichocarpa] |
| 20 |
Hb_001525_010 |
0.1907810187 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |