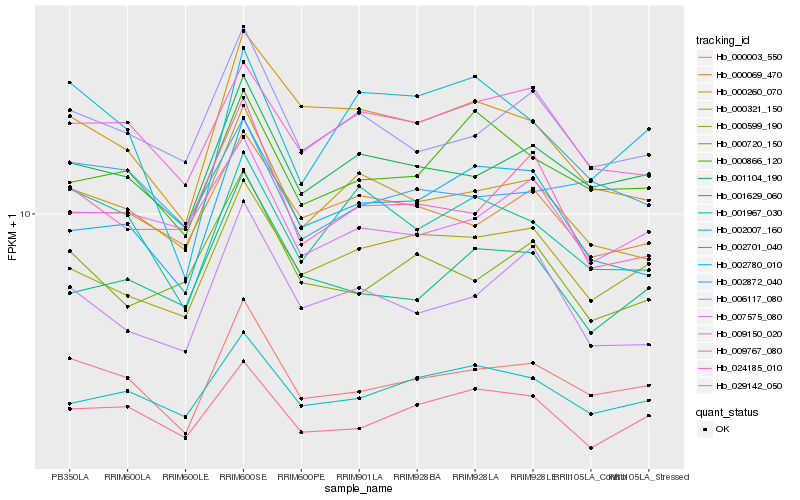
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_550 |
0.0 |
- |
- |
- |
| 2 |
Hb_001104_190 |
0.0799246756 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 3 |
Hb_006117_080 |
0.0942822192 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000321_150 |
0.0991612734 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 5 |
Hb_029142_050 |
0.1007853283 |
desease resistance |
Gene Name: PPR |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 6 |
Hb_002007_160 |
0.1017348805 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 7 |
Hb_000866_120 |
0.1037003443 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12 [Jatropha curcas] |
| 8 |
Hb_000599_190 |
0.1062525317 |
transcription factor |
TF Family: SNF2 |
DNA repair and recombination protein RAD26, putative [Ricinus communis] |
| 9 |
Hb_007575_080 |
0.1078219375 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 10 |
Hb_001629_060 |
0.1084874492 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
Flavin-containing amine oxidase domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002872_040 |
0.1087191985 |
- |
- |
pumilio, putative [Ricinus communis] |
| 12 |
Hb_001967_030 |
0.1105731741 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 13 |
Hb_002780_010 |
0.1113735722 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 14 |
Hb_024185_010 |
0.1124822993 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009150_020 |
0.1126373601 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 16 |
Hb_000069_470 |
0.1127629829 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 17 |
Hb_009767_080 |
0.1130230394 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 18 |
Hb_000260_070 |
0.1140314293 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_002701_040 |
0.114861714 |
- |
- |
PREDICTED: auxin transport protein BIG [Jatropha curcas] |
| 20 |
Hb_000720_150 |
0.1154077625 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X3 [Jatropha curcas] |