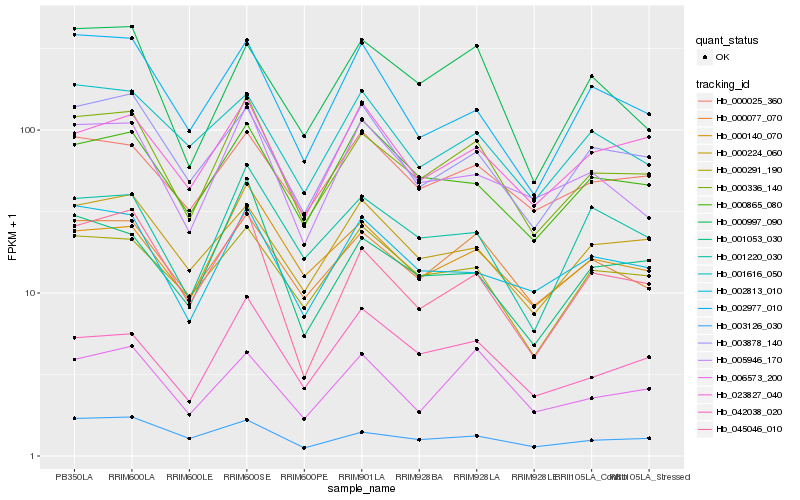
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000336_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000077_070 |
0.0767303957 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000140_070 |
0.0925512791 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 4 |
Hb_045046_010 |
0.0933237249 |
transcription factor |
TF Family: TUB |
hypothetical protein POPTR_0011s11070g [Populus trichocarpa] |
| 5 |
Hb_003126_030 |
0.0946555608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003878_140 |
0.0958145661 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 7 |
Hb_002813_010 |
0.0960407963 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001053_030 |
0.1014345706 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_005946_170 |
0.101582075 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 10 |
Hb_000224_060 |
0.1021472896 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 11 |
Hb_000291_190 |
0.1026433464 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 12 |
Hb_001220_030 |
0.102793 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 13 |
Hb_000865_080 |
0.1038045667 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 14 |
Hb_000025_360 |
0.1048637567 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006573_200 |
0.1051294442 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g52850, chloroplastic [Jatropha curcas] |
| 16 |
Hb_042038_020 |
0.1092208833 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 17 |
Hb_023827_040 |
0.1096713742 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 18 |
Hb_001616_050 |
0.1107324015 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 19 |
Hb_000997_090 |
0.1109789625 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 20 |
Hb_002977_010 |
0.1110904497 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |