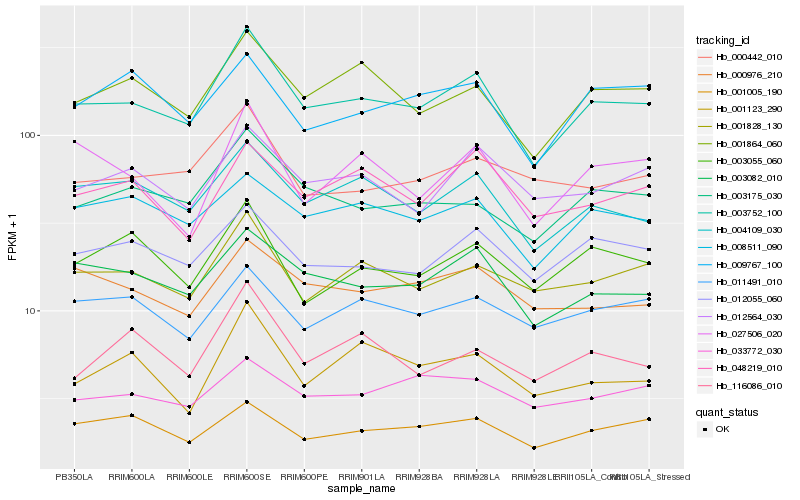
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003752_100 |
0.0 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 2 |
Hb_003082_010 |
0.0791318999 |
- |
- |
PREDICTED: putative F-box protein At3g10240 [Jatropha curcas] |
| 3 |
Hb_004109_030 |
0.0793620563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001864_060 |
0.0809527576 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 5 |
Hb_003175_030 |
0.084771981 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 6 |
Hb_001828_130 |
0.0867082827 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 7 |
Hb_012055_060 |
0.0904616681 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008511_090 |
0.0940341758 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 9 |
Hb_012564_030 |
0.0948120019 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 10 |
Hb_003055_060 |
0.0985599724 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105649487 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000442_010 |
0.0995533236 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 12 |
Hb_027506_020 |
0.0999985152 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 13 |
Hb_048219_010 |
0.1010599119 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 14 |
Hb_000976_210 |
0.1011971279 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_116086_010 |
0.1025659511 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 16 |
Hb_001005_190 |
0.1025854924 |
- |
- |
PREDICTED: UPF0496 protein At3g19330-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001123_290 |
0.1030343237 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 18 |
Hb_011491_010 |
0.1034328379 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 19 |
Hb_009767_100 |
0.1064189905 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 20 |
Hb_033772_030 |
0.1073617542 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |