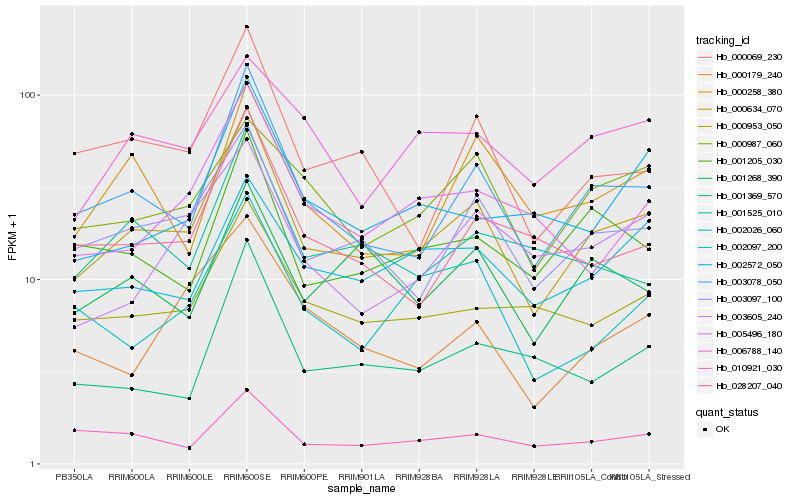
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000634_070 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 2 |
Hb_003078_050 |
0.0879540119 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 3 |
Hb_003097_100 |
0.101944546 |
- |
- |
probable glycerol-3-phosphate dehydrogenase [NAD(+)] 1, cytosolic [Jatropha curcas] |
| 4 |
Hb_000069_230 |
0.1264171816 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 5 |
Hb_000953_050 |
0.1348681707 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 6 |
Hb_001205_030 |
0.1414378776 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0007s12970g [Populus trichocarpa] |
| 7 |
Hb_028207_040 |
0.1428273355 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 8 |
Hb_002026_060 |
0.144524843 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 9 |
Hb_010921_030 |
0.1449449048 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At2g01810 [Jatropha curcas] |
| 10 |
Hb_005496_180 |
0.1460933909 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 11 |
Hb_002097_200 |
0.1478359363 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 12 |
Hb_001525_010 |
0.1492644826 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 13 |
Hb_001268_390 |
0.1497179001 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 14 |
Hb_000179_240 |
0.1506417124 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001369_570 |
0.1510211332 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 16 |
Hb_003605_240 |
0.1510638409 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 17 |
Hb_000987_060 |
0.151158067 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 18 |
Hb_000258_380 |
0.1523344484 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP11-like [Jatropha curcas] |
| 19 |
Hb_002572_050 |
0.152484826 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_006788_140 |
0.1548231804 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |