| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
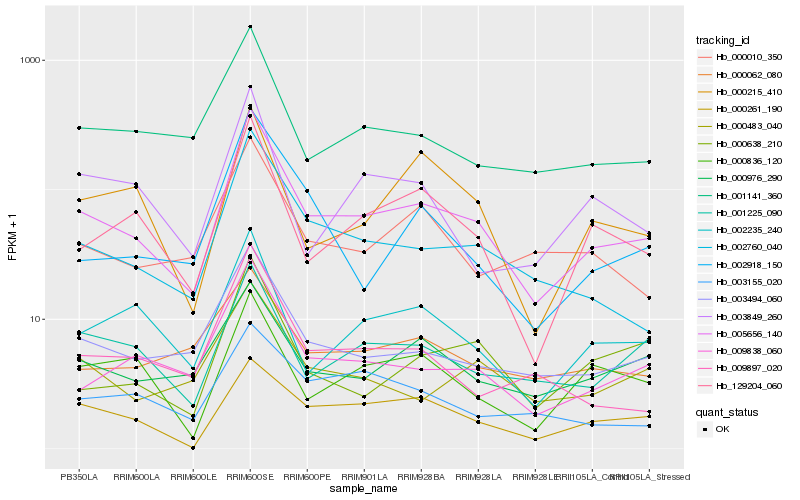
Hb_005656_140 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 2 |
Hb_003494_060 |
0.1291440633 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002760_040 |
0.1303014269 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 4 |
Hb_009838_060 |
0.1359490144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_129204_060 |
0.1414263744 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 6 |
Hb_001141_360 |
0.1472336621 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 7 |
Hb_002235_240 |
0.1492785832 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 8 |
Hb_001225_090 |
0.1508836889 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000976_290 |
0.1583332842 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 10 |
Hb_000261_190 |
0.1592200994 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Jatropha curcas] |
| 11 |
Hb_000483_040 |
0.1648390513 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |
| 12 |
Hb_000010_350 |
0.1650023877 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 13 |
Hb_009897_020 |
0.1677858123 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 14 |
Hb_000638_210 |
0.1693106487 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000836_120 |
0.1705145129 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 16 |
Hb_000062_080 |
0.1707518322 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 17 |
Hb_000215_410 |
0.1711833557 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 18 |
Hb_002918_150 |
0.1715664476 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 19 |
Hb_003849_260 |
0.1717728485 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 20 |
Hb_003155_020 |
0.1759713798 |
- |
- |
Rpp4C3 [Medicago truncatula] |