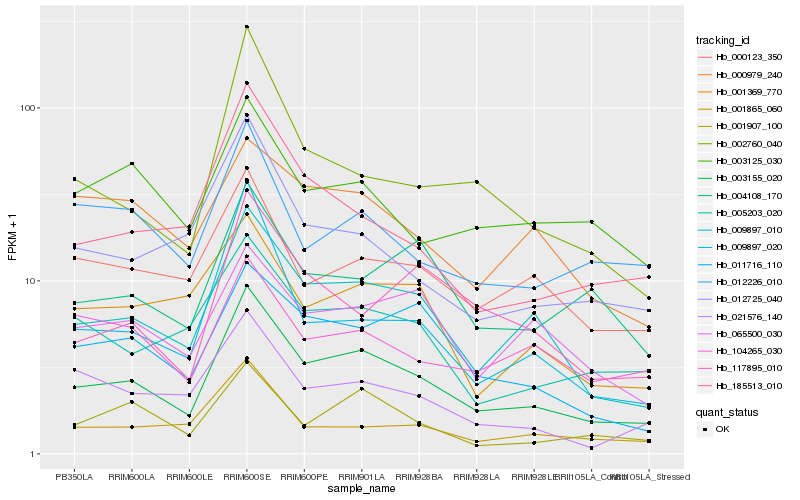
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003155_020 |
0.0 |
- |
- |
Rpp4C3 [Medicago truncatula] |
| 2 |
Hb_009897_010 |
0.0936614606 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 3 |
Hb_000123_350 |
0.1430178388 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_005203_020 |
0.1437977536 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 5 |
Hb_001865_060 |
0.144609844 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 6 |
Hb_011716_110 |
0.1450409674 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 7 |
Hb_001907_100 |
0.1485428922 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 8 |
Hb_012725_040 |
0.1510973734 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_004108_170 |
0.1523732736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001369_770 |
0.1535611138 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 11 |
Hb_185513_010 |
0.1537812733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_009897_020 |
0.1547137952 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 13 |
Hb_065500_030 |
0.1556107272 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 14 |
Hb_012226_010 |
0.1557826991 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 15 |
Hb_002760_040 |
0.1563410899 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 16 |
Hb_021576_140 |
0.1568558744 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 17 |
Hb_117895_010 |
0.156982955 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 18 |
Hb_003125_030 |
0.159462295 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 19 |
Hb_104265_030 |
0.1616304169 |
- |
- |
hypothetical protein POPTR_0001s31230g [Populus trichocarpa] |
| 20 |
Hb_000979_240 |
0.1617539268 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |