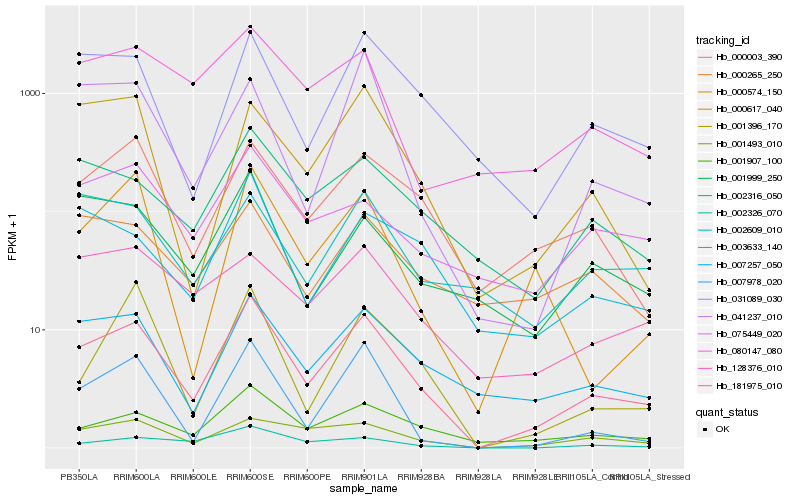
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_181975_010 |
0.0 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 2 |
Hb_007257_050 |
0.1525755712 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_001907_100 |
0.154254267 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 4 |
Hb_000003_390 |
0.1688233398 |
- |
- |
hypothetical protein POPTR_0003s20040g [Populus trichocarpa] |
| 5 |
Hb_031089_030 |
0.1694356245 |
- |
- |
ubiquitin-conjugating enzyme E2 28 [Arabidopsis thaliana] |
| 6 |
Hb_007978_020 |
0.1740486107 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA1-like [Jatropha curcas] |
| 7 |
Hb_001493_010 |
0.174502693 |
- |
- |
triacylglycerol acidic lipase TAL4 [Ricinus communis] |
| 8 |
Hb_002316_050 |
0.1803798389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002326_070 |
0.1871385183 |
- |
- |
ent-kaurene oxidase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003633_140 |
0.1884839187 |
- |
- |
PREDICTED: uncharacterized protein LOC105647486 isoform X1 [Jatropha curcas] |
| 11 |
Hb_080147_080 |
0.1886428362 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 12 |
Hb_001999_250 |
0.1896317083 |
- |
- |
PREDICTED: uncharacterized protein LOC105133073 isoform X1 [Populus euphratica] |
| 13 |
Hb_000617_040 |
0.1898503272 |
- |
- |
PREDICTED: uncharacterized protein LOC105647487 isoform X2 [Jatropha curcas] |
| 14 |
Hb_075449_020 |
0.1970177639 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 15 |
Hb_000574_150 |
0.1983235135 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 16 |
Hb_128376_010 |
0.1997403795 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000265_250 |
0.2013319143 |
- |
- |
auxilin, putative [Ricinus communis] |
| 18 |
Hb_001396_170 |
0.2021135291 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_041237_010 |
0.2026515318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002609_010 |
0.2032114676 |
- |
- |
PREDICTED: uncharacterized protein LOC105630866 [Jatropha curcas] |