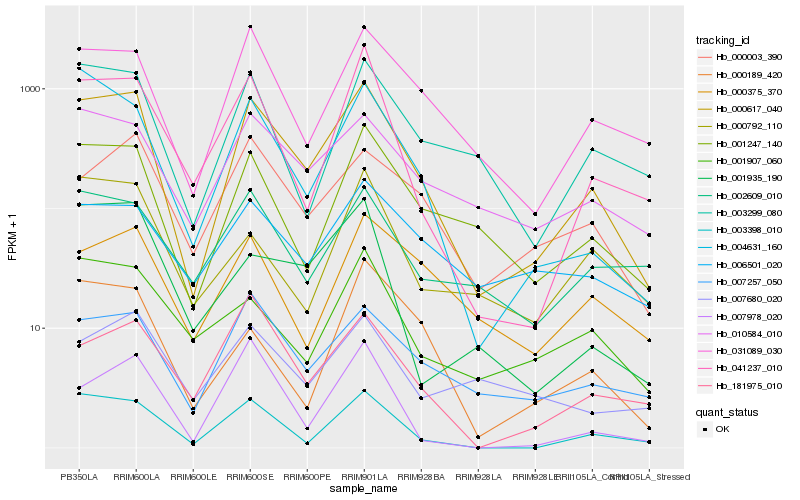
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000617_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647487 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001247_140 |
0.1493129542 |
- |
- |
- |
| 3 |
Hb_003398_010 |
0.15089202 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Eucalyptus grandis] |
| 4 |
Hb_031089_030 |
0.1569740232 |
- |
- |
ubiquitin-conjugating enzyme E2 28 [Arabidopsis thaliana] |
| 5 |
Hb_004631_160 |
0.1635032592 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 6 |
Hb_007257_050 |
0.1638441176 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_041237_010 |
0.1655462123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010584_010 |
0.1689405329 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 9 |
Hb_003299_080 |
0.1710119169 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 10 |
Hb_000003_390 |
0.1802431151 |
- |
- |
hypothetical protein POPTR_0003s20040g [Populus trichocarpa] |
| 11 |
Hb_181975_010 |
0.1898503272 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 12 |
Hb_001907_060 |
0.1967534076 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |
| 13 |
Hb_000375_370 |
0.1974417143 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_001935_190 |
0.1978838639 |
- |
- |
Nuc-1 negative regulatory protein preg, putative [Ricinus communis] |
| 15 |
Hb_002609_010 |
0.1979893107 |
- |
- |
PREDICTED: uncharacterized protein LOC105630866 [Jatropha curcas] |
| 16 |
Hb_000189_420 |
0.2012346361 |
- |
- |
PREDICTED: uncharacterized protein LOC105647665 [Jatropha curcas] |
| 17 |
Hb_006501_020 |
0.2013292391 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 18 |
Hb_007978_020 |
0.2052737148 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA1-like [Jatropha curcas] |
| 19 |
Hb_000792_110 |
0.2069786922 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 20 |
Hb_007680_020 |
0.2078450312 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |