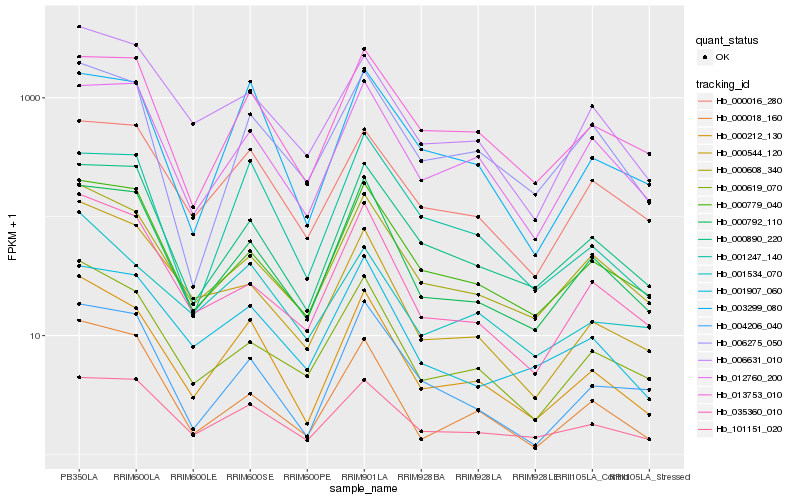
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000792_110 |
0.1214087221 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 3 |
Hb_000619_070 |
0.1243786338 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH1 [Jatropha curcas] |
| 4 |
Hb_101151_020 |
0.127996692 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000608_340 |
0.1285375988 |
- |
- |
PREDICTED: putative RNA-binding protein Luc7-like 2 [Populus euphratica] |
| 6 |
Hb_003299_080 |
0.1294756551 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 7 |
Hb_035360_010 |
0.1342142656 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 8 |
Hb_006631_010 |
0.1344031877 |
- |
- |
plasma membrane aquaporin 2 [Hevea brasiliensis] |
| 9 |
Hb_000779_040 |
0.1373580955 |
- |
- |
PREDICTED: protein FLX-like 4 [Jatropha curcas] |
| 10 |
Hb_000890_220 |
0.1375124744 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 11 |
Hb_001534_070 |
0.1381023254 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 12 |
Hb_000544_120 |
0.1382058818 |
- |
- |
PREDICTED: thymidylate kinase [Jatropha curcas] |
| 13 |
Hb_001907_060 |
0.1388847565 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |
| 14 |
Hb_000016_280 |
0.1416346331 |
- |
- |
Arginine/serine-rich splicing factor 35 [Theobroma cacao] |
| 15 |
Hb_006275_050 |
0.1416461869 |
- |
- |
CP3 [Hevea brasiliensis] |
| 16 |
Hb_004206_040 |
0.1424745729 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_000018_160 |
0.1426396055 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 18 |
Hb_013753_010 |
0.1446163746 |
- |
- |
PREDICTED: uncharacterized protein LOC105647969 isoform X1 [Jatropha curcas] |
| 19 |
Hb_012760_200 |
0.1469356285 |
- |
- |
protein phsophatase-2a, putative [Ricinus communis] |
| 20 |
Hb_001247_140 |
0.1490401923 |
- |
- |
- |