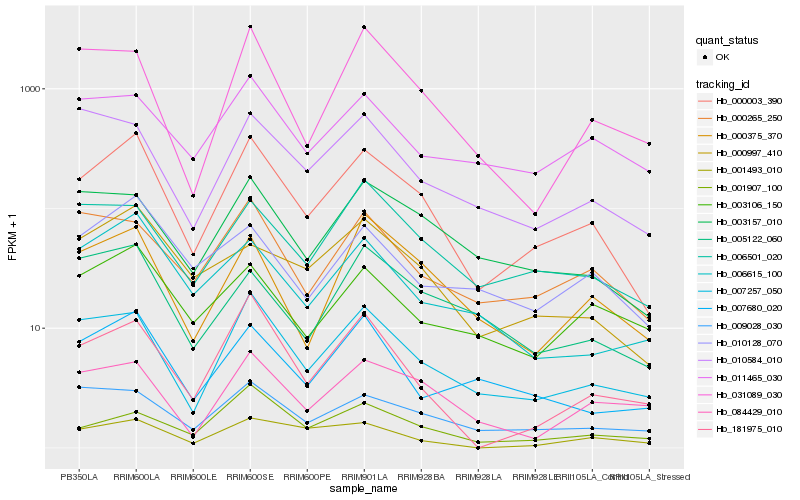
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_390 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s20040g [Populus trichocarpa] |
| 2 |
Hb_007257_050 |
0.1287634549 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_003157_010 |
0.1579259882 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 4 |
Hb_000997_410 |
0.1590650149 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 5 |
Hb_000375_370 |
0.1597708069 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_009028_030 |
0.1644597844 |
desease resistance |
Gene Name: NB-ARC |
NBS type disease resistance protein, putative [Theobroma cacao] |
| 7 |
Hb_006501_020 |
0.166019135 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 8 |
Hb_001907_100 |
0.1678384039 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 9 |
Hb_000265_250 |
0.1682985269 |
- |
- |
auxilin, putative [Ricinus communis] |
| 10 |
Hb_181975_010 |
0.1688233398 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 11 |
Hb_006615_100 |
0.1692677228 |
- |
- |
PREDICTED: uncharacterized protein LOC105640683 isoform X1 [Jatropha curcas] |
| 12 |
Hb_010128_070 |
0.1703853773 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_007680_020 |
0.1718145419 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003106_150 |
0.1758848672 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |
| 15 |
Hb_084429_010 |
0.1759751922 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Populus euphratica] |
| 16 |
Hb_031089_030 |
0.1775828609 |
- |
- |
ubiquitin-conjugating enzyme E2 28 [Arabidopsis thaliana] |
| 17 |
Hb_010584_010 |
0.1781367522 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 18 |
Hb_011465_030 |
0.1783351233 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 19 |
Hb_001493_010 |
0.1789893534 |
- |
- |
triacylglycerol acidic lipase TAL4 [Ricinus communis] |
| 20 |
Hb_005122_060 |
0.18023776 |
- |
- |
unnamed protein product [Vitis vinifera] |