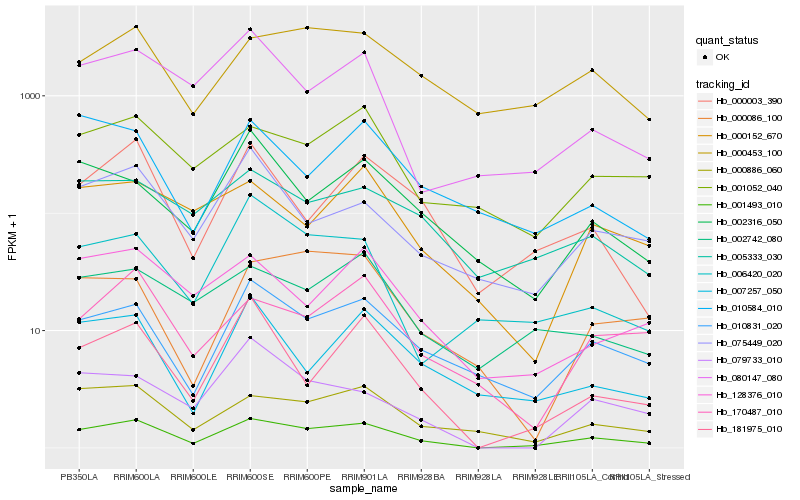
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001493_010 |
0.0 |
- |
- |
triacylglycerol acidic lipase TAL4 [Ricinus communis] |
| 2 |
Hb_010831_020 |
0.1477583685 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_000886_060 |
0.1613434878 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK10 isoform X2 [Vitis vinifera] |
| 4 |
Hb_181975_010 |
0.174502693 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 5 |
Hb_000003_390 |
0.1789893534 |
- |
- |
hypothetical protein POPTR_0003s20040g [Populus trichocarpa] |
| 6 |
Hb_170487_010 |
0.1790841203 |
- |
- |
- |
| 7 |
Hb_001052_040 |
0.1806738279 |
- |
- |
Ferredoxin-3, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_075449_020 |
0.1840020053 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 9 |
Hb_000453_100 |
0.1854760146 |
- |
- |
PREDICTED: metallothionein-like protein 1 [Sesamum indicum] |
| 10 |
Hb_002742_080 |
0.1867854554 |
- |
- |
unknown [Populus trichocarpa] |
| 11 |
Hb_128376_010 |
0.1868282522 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_005333_030 |
0.1886524672 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 13 |
Hb_080147_080 |
0.1913349067 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 14 |
Hb_007257_050 |
0.1919680844 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_079733_010 |
0.1924877596 |
- |
- |
- |
| 16 |
Hb_002316_050 |
0.1928034998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000086_100 |
0.1940296686 |
- |
- |
hypothetical protein POPTR_0005s24730g [Populus trichocarpa] |
| 18 |
Hb_000152_670 |
0.1964167284 |
- |
- |
hypothetical protein VITISV_037603 [Vitis vinifera] |
| 19 |
Hb_006420_020 |
0.1974117622 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 20 |
Hb_010584_010 |
0.200563412 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |