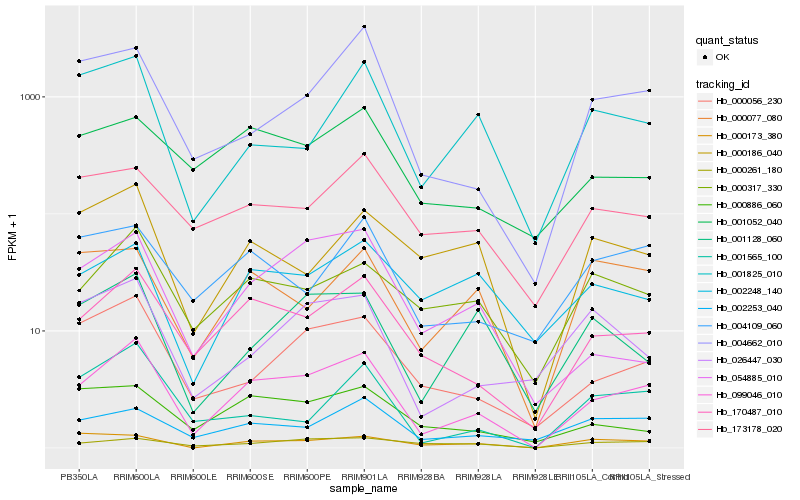
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_099046_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 2 |
Hb_170487_010 |
0.1432935504 |
- |
- |
- |
| 3 |
Hb_000056_230 |
0.1777580981 |
transcription factor |
TF Family: TCP |
hypothetical protein JCGZ_15588 [Jatropha curcas] |
| 4 |
Hb_000317_330 |
0.19144802 |
- |
- |
PREDICTED: RNA-binding protein 38-like [Jatropha curcas] |
| 5 |
Hb_004662_010 |
0.2018747728 |
- |
- |
Nascent polypeptide-associated complex subunit beta [Medicago truncatula] |
| 6 |
Hb_002253_040 |
0.2035011789 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 7 |
Hb_001565_100 |
0.205917462 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_000261_180 |
0.2067581659 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_001052_040 |
0.2097815774 |
- |
- |
Ferredoxin-3, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_004109_060 |
0.211707756 |
- |
- |
hypothetical protein POPTR_0019s12520g [Populus trichocarpa] |
| 11 |
Hb_173178_020 |
0.2130239822 |
- |
- |
PREDICTED: vesicle transport v-SNARE 12-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000186_040 |
0.2148676021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_026447_030 |
0.2158315539 |
- |
- |
hypothetical protein POPTR_0006s06500g [Populus trichocarpa] |
| 14 |
Hb_001128_060 |
0.216313866 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 15 |
Hb_001825_010 |
0.2163225403 |
- |
- |
PREDICTED: bet1-like SNARE 1-1 [Jatropha curcas] |
| 16 |
Hb_000886_060 |
0.2167216671 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK10 isoform X2 [Vitis vinifera] |
| 17 |
Hb_000077_080 |
0.2173079373 |
- |
- |
PREDICTED: uncharacterized protein LOC105646929 [Jatropha curcas] |
| 18 |
Hb_000173_380 |
0.2184447709 |
transcription factor |
TF Family: FAR1 |
- |
| 19 |
Hb_002248_140 |
0.21863732 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 20 |
Hb_054885_010 |
0.2214069875 |
- |
- |
- |