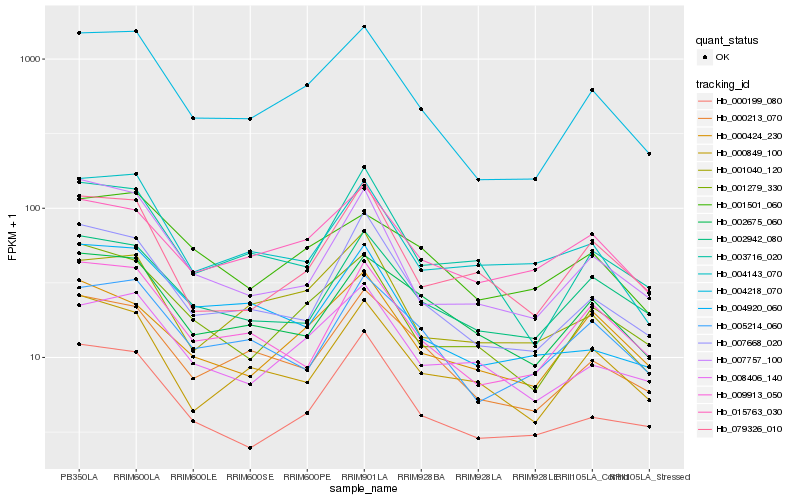
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_070 |
0.0 |
- |
- |
annexin [Manihot esculenta] |
| 2 |
Hb_001279_330 |
0.093871326 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 3 |
Hb_007668_020 |
0.0956536491 |
- |
- |
PREDICTED: uncharacterized protein LOC105631102 [Jatropha curcas] |
| 4 |
Hb_000213_070 |
0.0995961323 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |
| 5 |
Hb_009913_050 |
0.1084167192 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 6 |
Hb_007757_100 |
0.1088650729 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |
| 7 |
Hb_000199_080 |
0.1112101433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008406_140 |
0.1128444667 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 9 |
Hb_005214_060 |
0.1142165515 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_001040_120 |
0.1143159307 |
- |
- |
allantoinase, putative [Ricinus communis] |
| 11 |
Hb_003716_020 |
0.1145465556 |
- |
- |
PREDICTED: putative vesicle-associated membrane protein 726 [Jatropha curcas] |
| 12 |
Hb_000424_230 |
0.1168011622 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Glycine max] |
| 13 |
Hb_000849_100 |
0.1170889377 |
- |
- |
PREDICTED: uncharacterized protein LOC105644134 [Jatropha curcas] |
| 14 |
Hb_002942_080 |
0.1174798549 |
- |
- |
PREDICTED: signal recognition particle subunit SRP68 [Jatropha curcas] |
| 15 |
Hb_001501_060 |
0.1200291319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002675_060 |
0.1202316837 |
- |
- |
PREDICTED: uncharacterized protein LOC105634954 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004920_060 |
0.123047558 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 18 |
Hb_015763_030 |
0.1245136629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004143_070 |
0.1249244504 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_079326_010 |
0.1250949771 |
- |
- |
PREDICTED: uncharacterized protein LOC105630719 isoform X1 [Jatropha curcas] |