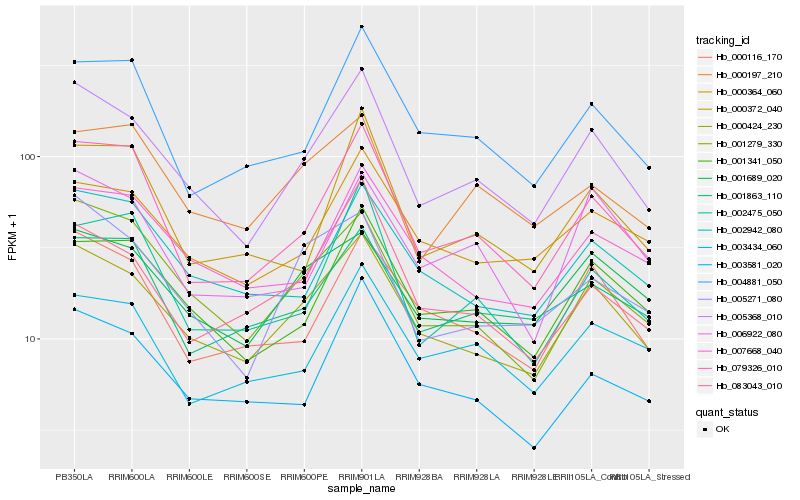
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005368_010 |
0.0 |
- |
- |
PREDICTED: umecyanin-like [Jatropha curcas] |
| 2 |
Hb_000424_230 |
0.0620341313 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Glycine max] |
| 3 |
Hb_079326_010 |
0.0831682196 |
- |
- |
PREDICTED: uncharacterized protein LOC105630719 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001341_050 |
0.090706967 |
- |
- |
ATP synthase D chain, mitochondrial [Theobroma cacao] |
| 5 |
Hb_001279_330 |
0.0982782562 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 6 |
Hb_005271_080 |
0.1047681087 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 7 |
Hb_000116_170 |
0.1060305464 |
- |
- |
hypothetical protein JCGZ_22392 [Jatropha curcas] |
| 8 |
Hb_004881_050 |
0.1082583927 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 9 |
Hb_002942_080 |
0.1113557329 |
- |
- |
PREDICTED: signal recognition particle subunit SRP68 [Jatropha curcas] |
| 10 |
Hb_000364_060 |
0.1115309514 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 11 |
Hb_001689_020 |
0.1126804089 |
- |
- |
hypothetical protein JCGZ_10660 [Jatropha curcas] |
| 12 |
Hb_083043_010 |
0.1128517867 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 13 |
Hb_003581_020 |
0.1130910755 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000372_040 |
0.1138828285 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_007668_040 |
0.1146846653 |
- |
- |
PREDICTED: uncharacterized protein LOC103409082 [Malus domestica] |
| 16 |
Hb_002475_050 |
0.1160638569 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 17 |
Hb_001863_110 |
0.1177013821 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006922_080 |
0.1195148816 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT3-like [Glycine max] |
| 19 |
Hb_003434_060 |
0.1198332685 |
- |
- |
PREDICTED: membrane-bound transcription factor site-2 protease homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_000197_210 |
0.1200790104 |
- |
- |
phosphatidylinositol transporter, putative [Ricinus communis] |