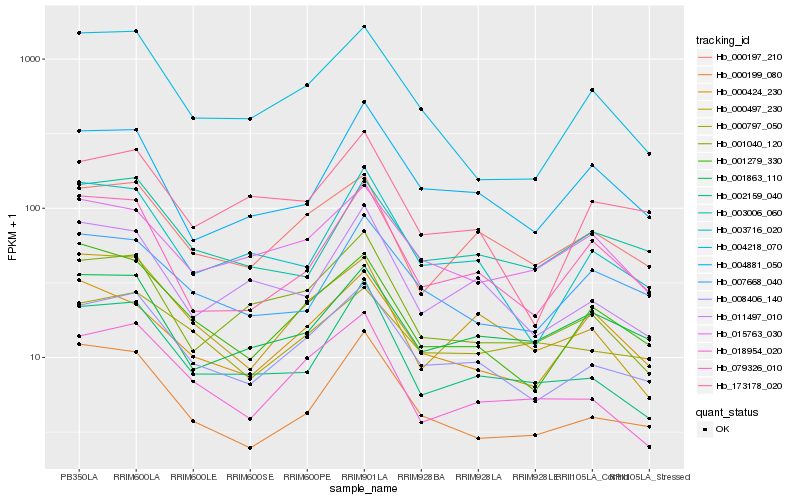
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008406_140 |
0.0 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 2 |
Hb_000197_210 |
0.0903519065 |
- |
- |
phosphatidylinositol transporter, putative [Ricinus communis] |
| 3 |
Hb_018954_020 |
0.1001540273 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH13 [Jatropha curcas] |
| 4 |
Hb_001279_330 |
0.1011159062 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 5 |
Hb_000199_080 |
0.1020327813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002159_040 |
0.1041045183 |
- |
- |
hypothetical protein PRUPE_ppa001563mg [Prunus persica] |
| 7 |
Hb_003716_020 |
0.1069206432 |
- |
- |
PREDICTED: putative vesicle-associated membrane protein 726 [Jatropha curcas] |
| 8 |
Hb_001040_120 |
0.1084274823 |
- |
- |
allantoinase, putative [Ricinus communis] |
| 9 |
Hb_079326_010 |
0.1085404334 |
- |
- |
PREDICTED: uncharacterized protein LOC105630719 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000497_230 |
0.1088917932 |
- |
- |
PREDICTED: probable protein S-acyltransferase 6 [Jatropha curcas] |
| 11 |
Hb_000797_050 |
0.1108011047 |
- |
- |
PREDICTED: uncharacterized protein LOC105645916 [Jatropha curcas] |
| 12 |
Hb_011497_010 |
0.1112393157 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 13 |
Hb_001863_110 |
0.1128402338 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004218_070 |
0.1128444667 |
- |
- |
annexin [Manihot esculenta] |
| 15 |
Hb_003006_060 |
0.1136745494 |
- |
- |
PREDICTED: pre-mRNA-splicing factor syf2 [Jatropha curcas] |
| 16 |
Hb_004881_050 |
0.1137938983 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 17 |
Hb_015763_030 |
0.114433232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_173178_020 |
0.1159303232 |
- |
- |
PREDICTED: vesicle transport v-SNARE 12-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_007668_040 |
0.1163492042 |
- |
- |
PREDICTED: uncharacterized protein LOC103409082 [Malus domestica] |
| 20 |
Hb_000424_230 |
0.1177625684 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Glycine max] |