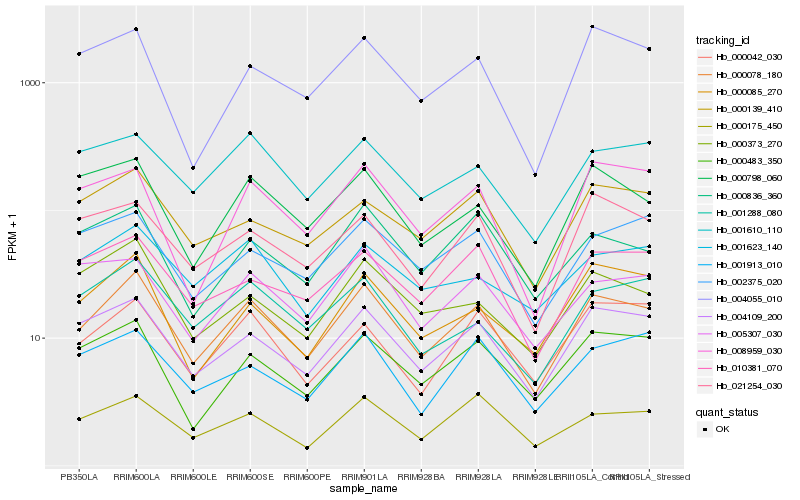
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000078_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641312 [Jatropha curcas] |
| 2 |
Hb_004109_200 |
0.0857117581 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000175_450 |
0.108879187 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g52620 [Jatropha curcas] |
| 4 |
Hb_000483_350 |
0.1105307482 |
- |
- |
PREDICTED: uncharacterized protein LOC105632381 [Jatropha curcas] |
| 5 |
Hb_000798_060 |
0.1134923331 |
transcription factor |
TF Family: PHD |
- |
| 6 |
Hb_000085_270 |
0.1157568187 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 7 |
Hb_001288_080 |
0.1159692099 |
- |
- |
PREDICTED: probable protein phosphatase 2C 50 [Populus euphratica] |
| 8 |
Hb_001610_110 |
0.1162979364 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 32 homolog 2 [Jatropha curcas] |
| 9 |
Hb_010381_070 |
0.1166725359 |
- |
- |
PREDICTED: protein STU1-like [Jatropha curcas] |
| 10 |
Hb_000836_360 |
0.1174244461 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004055_010 |
0.1179549628 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 12 |
Hb_008959_030 |
0.1180344609 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |
| 13 |
Hb_000139_410 |
0.118313328 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 14 |
Hb_000042_030 |
0.118925109 |
transcription factor |
TF Family: NAC |
NAC domain protein, IPR003441 [Theobroma cacao] |
| 15 |
Hb_021254_030 |
0.1196583403 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein P [Jatropha curcas] |
| 16 |
Hb_000373_270 |
0.1208547013 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005307_030 |
0.1224436081 |
- |
- |
hypothetical protein POPTR_0006s04640g [Populus trichocarpa] |
| 18 |
Hb_001623_140 |
0.1228730894 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 19 |
Hb_002375_020 |
0.1234504501 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 65-like [Jatropha curcas] |
| 20 |
Hb_001913_010 |
0.123913406 |
- |
- |
phosphatidylinositol n-acetylglucosaminyltransferase subunit p, putative [Ricinus communis] |