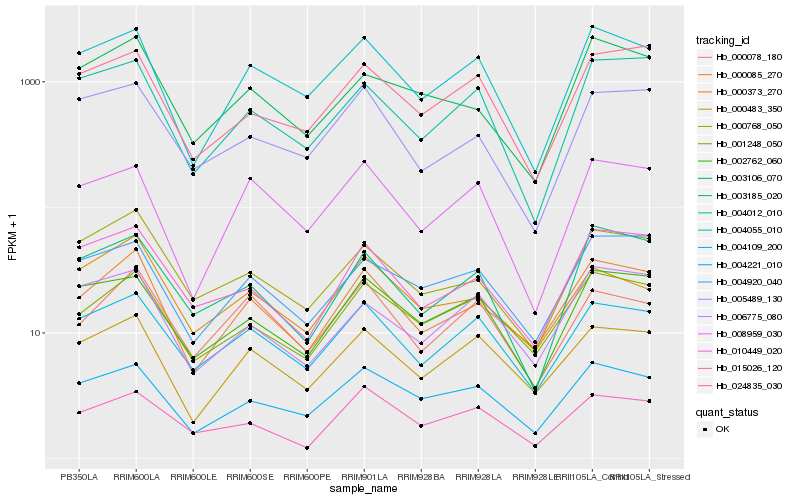
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000085_270 |
0.0 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 2 |
Hb_001248_050 |
0.0829095401 |
- |
- |
PREDICTED: PXMP2/4 family protein 4-like [Jatropha curcas] |
| 3 |
Hb_000483_350 |
0.0841226903 |
- |
- |
PREDICTED: uncharacterized protein LOC105632381 [Jatropha curcas] |
| 4 |
Hb_004109_200 |
0.0948388564 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_004221_010 |
0.0961117428 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 19-like [Jatropha curcas] |
| 6 |
Hb_004055_010 |
0.0977530418 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 7 |
Hb_004920_040 |
0.0992321112 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 8 |
Hb_010449_020 |
0.1009940594 |
- |
- |
PREDICTED: rRNA-processing protein UTP23 homolog [Jatropha curcas] |
| 9 |
Hb_000768_050 |
0.1048398357 |
- |
- |
hypothetical protein RCOM_0845740 [Ricinus communis] |
| 10 |
Hb_003106_070 |
0.1103992021 |
- |
- |
PREDICTED: uncharacterized protein LOC105167687 [Sesamum indicum] |
| 11 |
Hb_002762_060 |
0.112832403 |
- |
- |
PREDICTED: diphthine methyltransferase homolog [Jatropha curcas] |
| 12 |
Hb_006775_080 |
0.1132606551 |
- |
- |
PREDICTED: uncharacterized protein LOC105649730 [Jatropha curcas] |
| 13 |
Hb_024835_030 |
0.113706767 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 14 |
Hb_000078_180 |
0.1157568187 |
- |
- |
PREDICTED: uncharacterized protein LOC105641312 [Jatropha curcas] |
| 15 |
Hb_003185_020 |
0.1166767829 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 50 [Jatropha curcas] |
| 16 |
Hb_000373_270 |
0.1180139032 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005489_130 |
0.1188628513 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 18 |
Hb_004012_010 |
0.1191746328 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1-like [Gossypium raimondii] |
| 19 |
Hb_015026_120 |
0.1196027663 |
- |
- |
hypothetical protein TRIUR3_03502 [Triticum urartu] |
| 20 |
Hb_008959_030 |
0.1204745664 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |