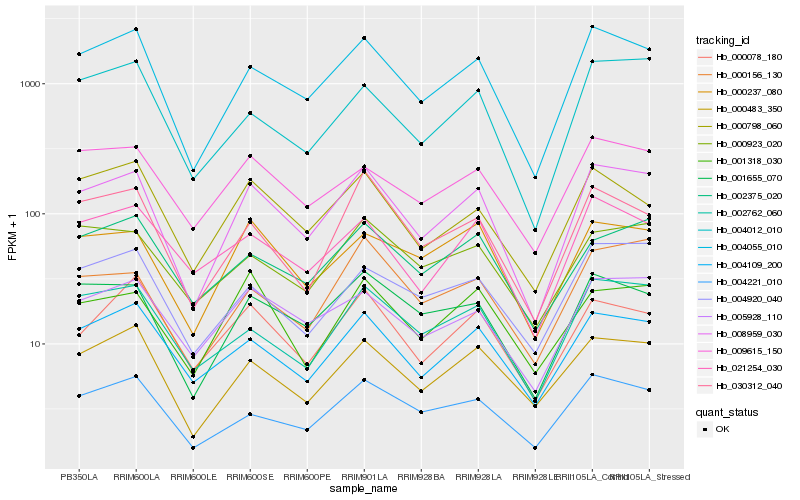
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008959_030 |
0.0 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |
| 2 |
Hb_004055_010 |
0.0592325025 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 3 |
Hb_001655_070 |
0.0875218608 |
- |
- |
PREDICTED: chromatin structure-remodeling complex subunit RSC1 [Jatropha curcas] |
| 4 |
Hb_000483_350 |
0.0948134644 |
- |
- |
PREDICTED: uncharacterized protein LOC105632381 [Jatropha curcas] |
| 5 |
Hb_004109_200 |
0.0974954504 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_005928_110 |
0.0982408045 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 7 |
Hb_004221_010 |
0.1022490567 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 19-like [Jatropha curcas] |
| 8 |
Hb_002762_060 |
0.1026001729 |
- |
- |
PREDICTED: diphthine methyltransferase homolog [Jatropha curcas] |
| 9 |
Hb_000237_080 |
0.1040717539 |
transcription factor |
TF Family: PHD |
PREDICTED: chromatin structure-remodeling complex subunit RSC1 [Jatropha curcas] |
| 10 |
Hb_030312_040 |
0.1081448323 |
- |
- |
hypothetical protein JCGZ_04876 [Jatropha curcas] |
| 11 |
Hb_004012_010 |
0.1086538951 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1-like [Gossypium raimondii] |
| 12 |
Hb_004920_040 |
0.1092241235 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 13 |
Hb_009615_150 |
0.1099561774 |
- |
- |
PREDICTED: transcription elongation factor SPT4 homolog 2 [Jatropha curcas] |
| 14 |
Hb_000156_130 |
0.1100368825 |
- |
- |
hypothetical protein JCGZ_04605 [Jatropha curcas] |
| 15 |
Hb_000798_060 |
0.112602483 |
transcription factor |
TF Family: PHD |
- |
| 16 |
Hb_021254_030 |
0.1128782951 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein P [Jatropha curcas] |
| 17 |
Hb_001318_030 |
0.1143942181 |
- |
- |
PREDICTED: uncharacterized protein LOC105642686 [Jatropha curcas] |
| 18 |
Hb_002375_020 |
0.1162867818 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 65-like [Jatropha curcas] |
| 19 |
Hb_000923_020 |
0.1174903184 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 65-like [Jatropha curcas] |
| 20 |
Hb_000078_180 |
0.1180344609 |
- |
- |
PREDICTED: uncharacterized protein LOC105641312 [Jatropha curcas] |