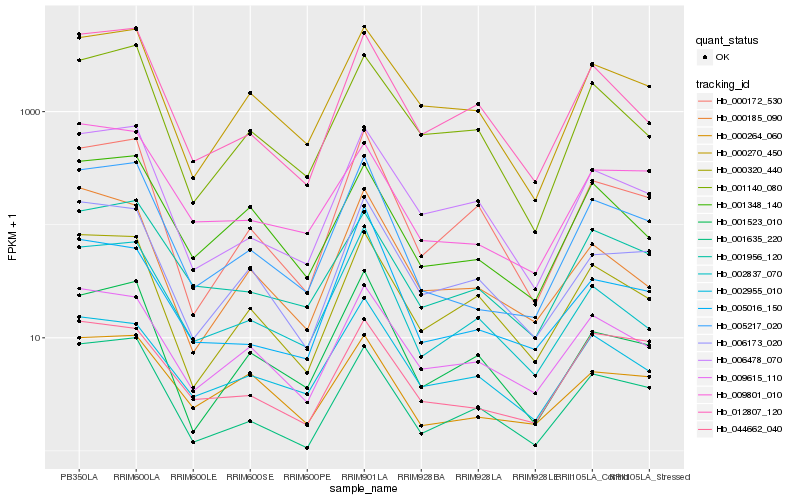
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005217_020 |
0.0 |
- |
- |
PREDICTED: ribosomal RNA processing protein 36 homolog isoform X2 [Jatropha curcas] |
| 2 |
Hb_000264_060 |
0.1161865311 |
- |
- |
PREDICTED: RNA polymerase-associated protein RTF1 homolog [Jatropha curcas] |
| 3 |
Hb_002837_070 |
0.1228818484 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 8-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000270_450 |
0.1266926089 |
- |
- |
PREDICTED: uncharacterized protein LOC105167687 [Sesamum indicum] |
| 5 |
Hb_001523_010 |
0.1277467884 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_25496 [Jatropha curcas] |
| 6 |
Hb_000172_530 |
0.1277713517 |
- |
- |
PREDICTED: UBP1-associated protein 2B-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_009615_110 |
0.1289540214 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 8 |
Hb_006478_070 |
0.1297038948 |
- |
- |
PREDICTED: uncharacterized protein LOC105649957 [Jatropha curcas] |
| 9 |
Hb_009801_010 |
0.1299314056 |
- |
- |
ATPase inhibitor, putative [Ricinus communis] |
| 10 |
Hb_001956_120 |
0.1305507574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001348_140 |
0.1306896333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006173_020 |
0.1311590326 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_044662_040 |
0.1368686163 |
- |
- |
- |
| 14 |
Hb_012807_120 |
0.1375782657 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_001635_220 |
0.1407364617 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 16 |
Hb_002955_010 |
0.1408783595 |
- |
- |
PREDICTED: Werner syndrome ATP-dependent helicase homolog [Jatropha curcas] |
| 17 |
Hb_000185_090 |
0.1410305504 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 18 |
Hb_000320_440 |
0.1410680016 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 2/3/4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001140_080 |
0.1412474948 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 20 |
Hb_005016_150 |
0.1417912515 |
- |
- |
- |