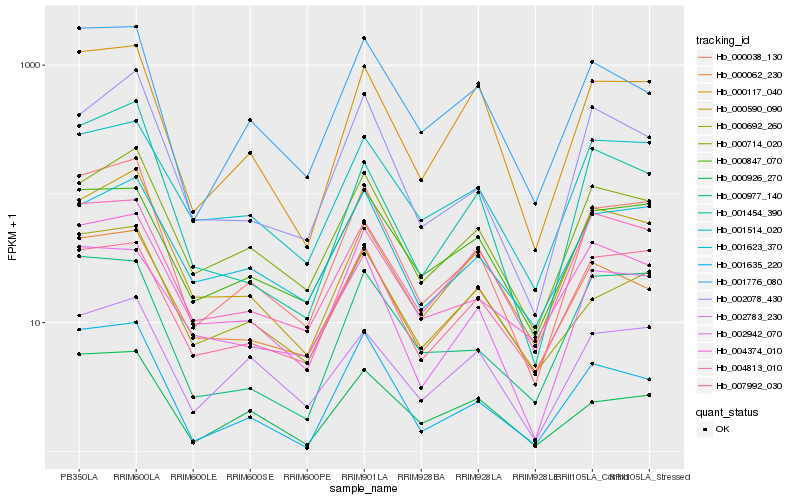
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000038_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_000714_020 |
0.103760961 |
- |
- |
PREDICTED: uncharacterized protein LOC105635336 [Jatropha curcas] |
| 3 |
Hb_001635_220 |
0.1098523946 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 4 |
Hb_000117_040 |
0.1111570637 |
- |
- |
30S ribosomal protein S6A [Hevea brasiliensis] |
| 5 |
Hb_002078_430 |
0.1138517826 |
- |
- |
hypothetical protein PRUPE_ppa014280mg [Prunus persica] |
| 6 |
Hb_000590_090 |
0.1144023244 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004374_010 |
0.1173261125 |
- |
- |
PREDICTED: PIN2/TERF1-interacting telomerase inhibitor 1 [Jatropha curcas] |
| 8 |
Hb_000692_260 |
0.117533596 |
- |
- |
PREDICTED: uncharacterized protein LOC105635282 [Jatropha curcas] |
| 9 |
Hb_002942_070 |
0.1195056327 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |
| 10 |
Hb_001454_390 |
0.120220536 |
- |
- |
8-kd dynein light chain, DLC8, pin [Thalassiosira pseudonana CCMP1335] |
| 11 |
Hb_000977_140 |
0.1202830468 |
- |
- |
PREDICTED: uncharacterized protein LOC105801335 [Gossypium raimondii] |
| 12 |
Hb_000926_270 |
0.1208210343 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 13 |
Hb_001623_370 |
0.1212062381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000062_230 |
0.1215031024 |
- |
- |
PREDICTED: probable thimet oligopeptidase isoform X1 [Jatropha curcas] |
| 15 |
Hb_001776_080 |
0.1217981747 |
- |
- |
PREDICTED: CAX-interacting protein 4 [Jatropha curcas] |
| 16 |
Hb_007992_030 |
0.1220080704 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105646951 [Jatropha curcas] |
| 17 |
Hb_001514_020 |
0.1222498435 |
- |
- |
PREDICTED: protein DCL, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_004813_010 |
0.1223302664 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_002783_230 |
0.1227121476 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 20 |
Hb_000847_070 |
0.1248033883 |
- |
- |
PREDICTED: uncharacterized protein LOC105125699 [Populus euphratica] |