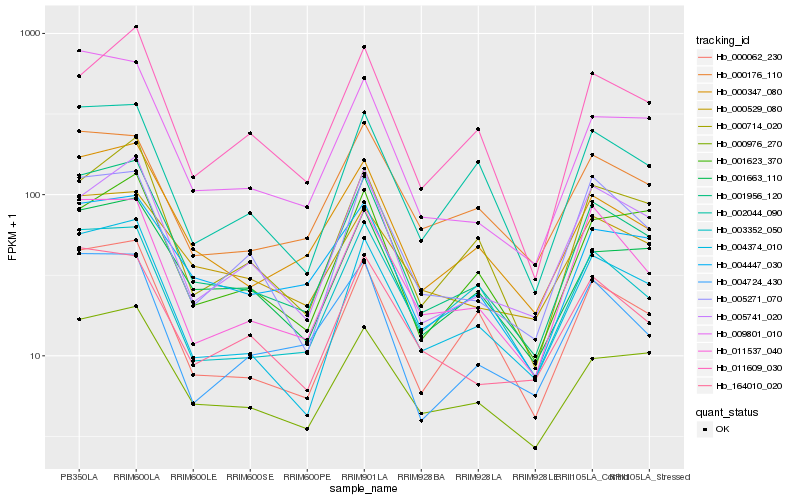
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001956_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004374_010 |
0.0689371143 |
- |
- |
PREDICTED: PIN2/TERF1-interacting telomerase inhibitor 1 [Jatropha curcas] |
| 3 |
Hb_000347_080 |
0.0702532661 |
- |
- |
PREDICTED: 25.3 kDa vesicle transport protein [Jatropha curcas] |
| 4 |
Hb_000976_270 |
0.0808107667 |
- |
- |
PREDICTED: uncharacterized protein LOC105646226 [Jatropha curcas] |
| 5 |
Hb_011537_040 |
0.085258716 |
- |
- |
PREDICTED: uncharacterized protein LOC105646229 [Jatropha curcas] |
| 6 |
Hb_000062_230 |
0.0853147476 |
- |
- |
PREDICTED: probable thimet oligopeptidase isoform X1 [Jatropha curcas] |
| 7 |
Hb_001663_110 |
0.0867536796 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_009801_010 |
0.0906979797 |
- |
- |
ATPase inhibitor, putative [Ricinus communis] |
| 9 |
Hb_000714_020 |
0.093307765 |
- |
- |
PREDICTED: uncharacterized protein LOC105635336 [Jatropha curcas] |
| 10 |
Hb_004447_030 |
0.0944561185 |
- |
- |
PREDICTED: glucose-induced degradation protein 4 homolog [Jatropha curcas] |
| 11 |
Hb_005741_020 |
0.094514447 |
- |
- |
PREDICTED: putative golgin subfamily A member 6-like protein 6 [Jatropha curcas] |
| 12 |
Hb_164010_020 |
0.0982667173 |
- |
- |
PREDICTED: DNA-directed RNA polymerases I and III subunit RPAC1 [Jatropha curcas] |
| 13 |
Hb_011609_030 |
0.0987229529 |
- |
- |
conserved hypothetical protein 13 [Hevea brasiliensis] |
| 14 |
Hb_005271_070 |
0.1001177218 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 15 |
Hb_002044_090 |
0.1014919539 |
- |
- |
PREDICTED: uncharacterized protein LOC105643541 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004724_430 |
0.1029435469 |
- |
- |
PREDICTED: F-box protein At2g27310 [Jatropha curcas] |
| 17 |
Hb_001623_370 |
0.1031941702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000176_110 |
0.1060982668 |
- |
- |
PREDICTED: serine--tRNA ligase [Jatropha curcas] |
| 19 |
Hb_003352_050 |
0.1079241535 |
- |
- |
PREDICTED: uncharacterized protein LOC105643709 [Jatropha curcas] |
| 20 |
Hb_000529_080 |
0.1082602363 |
- |
- |
PREDICTED: uncharacterized protein LOC105641675 [Jatropha curcas] |