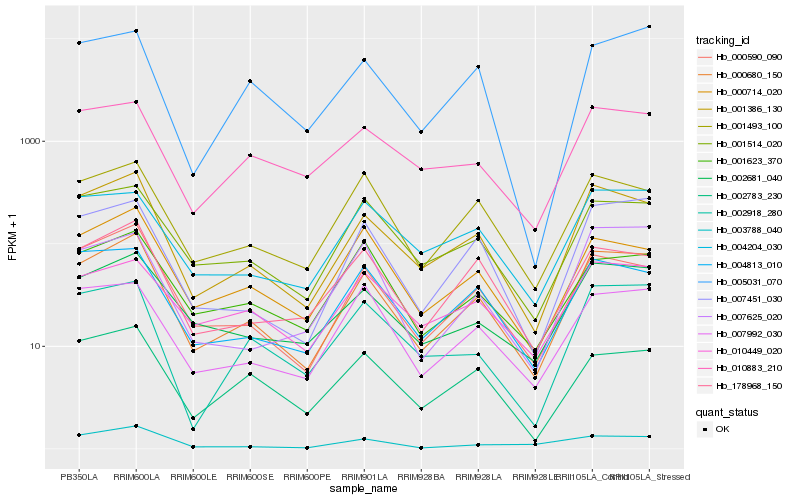
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000680_150 |
0.0 |
- |
- |
PREDICTED: protein STU1-like [Jatropha curcas] |
| 2 |
Hb_001386_130 |
0.0635583697 |
- |
- |
Mitochondrial import receptor subunit TOM20, putative [Ricinus communis] |
| 3 |
Hb_000590_090 |
0.0926565002 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_007451_030 |
0.1023390342 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |
| 5 |
Hb_002681_040 |
0.1078347325 |
- |
- |
PREDICTED: ribosomal RNA-processing protein 8 [Jatropha curcas] |
| 6 |
Hb_001514_020 |
0.1157767332 |
- |
- |
PREDICTED: protein DCL, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_001493_100 |
0.1185772081 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 34-like [Jatropha curcas] |
| 8 |
Hb_004813_010 |
0.1203543903 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_178968_150 |
0.1206879091 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000714_020 |
0.1239965192 |
- |
- |
PREDICTED: uncharacterized protein LOC105635336 [Jatropha curcas] |
| 11 |
Hb_002783_230 |
0.1254399949 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 12 |
Hb_007992_030 |
0.1255176015 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105646951 [Jatropha curcas] |
| 13 |
Hb_004204_030 |
0.1292562702 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_002918_280 |
0.1293624144 |
- |
- |
hypothetical protein POPTR_0004s04800g [Populus trichocarpa] |
| 15 |
Hb_003788_040 |
0.1319908854 |
- |
- |
- |
| 16 |
Hb_005031_070 |
0.1321026371 |
- |
- |
RecName: Full=Major latex allergen Hev b 5; AltName: Allergen=Hev b 5 [Hevea brasiliensis] |
| 17 |
Hb_010883_210 |
0.132500163 |
- |
- |
PREDICTED: selenoprotein K [Jatropha curcas] |
| 18 |
Hb_010449_020 |
0.1335229966 |
- |
- |
PREDICTED: rRNA-processing protein UTP23 homolog [Jatropha curcas] |
| 19 |
Hb_007625_020 |
0.1337259968 |
- |
- |
PREDICTED: casein kinase II subunit alpha-like [Sesamum indicum] |
| 20 |
Hb_001623_370 |
0.1351100712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |