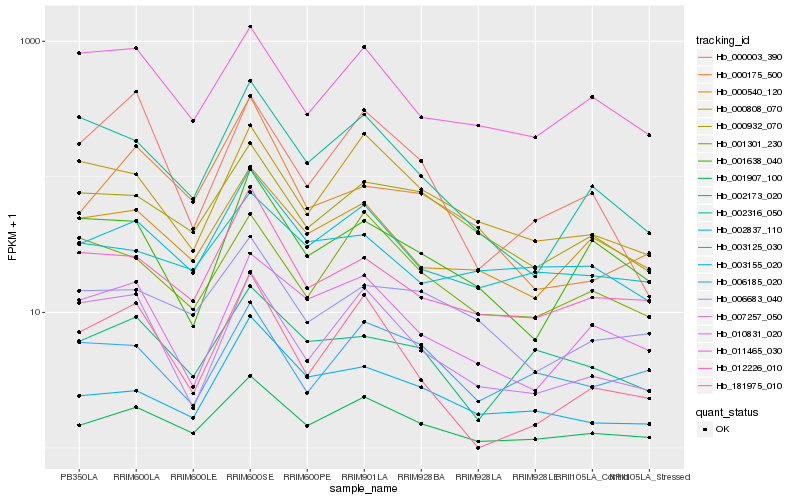
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001907_100 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 2 |
Hb_002316_050 |
0.144544271 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003155_020 |
0.1485428922 |
- |
- |
Rpp4C3 [Medicago truncatula] |
| 4 |
Hb_010831_020 |
0.1499620914 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 5 |
Hb_000808_070 |
0.1521356507 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |
| 6 |
Hb_181975_010 |
0.154254267 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 7 |
Hb_007257_050 |
0.1543269233 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000540_120 |
0.1556049456 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 9 |
Hb_012226_010 |
0.1561127057 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 10 |
Hb_000932_070 |
0.1642480902 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 11 |
Hb_000003_390 |
0.1678384039 |
- |
- |
hypothetical protein POPTR_0003s20040g [Populus trichocarpa] |
| 12 |
Hb_001301_230 |
0.169360947 |
- |
- |
PREDICTED: uncharacterized protein LOC105632949 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001638_040 |
0.169728886 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 14 |
Hb_000175_500 |
0.171449765 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 15 |
Hb_002173_020 |
0.1728676582 |
- |
- |
PREDICTED: uncharacterized protein LOC105801078 [Gossypium raimondii] |
| 16 |
Hb_002837_110 |
0.1743683517 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 2, mitochondrial [Jatropha curcas] |
| 17 |
Hb_006683_040 |
0.1748041668 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 18 |
Hb_011465_030 |
0.1749190399 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 19 |
Hb_003125_030 |
0.1755208536 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 20 |
Hb_006185_020 |
0.1766675918 |
- |
- |
PREDICTED: midasin [Jatropha curcas] |