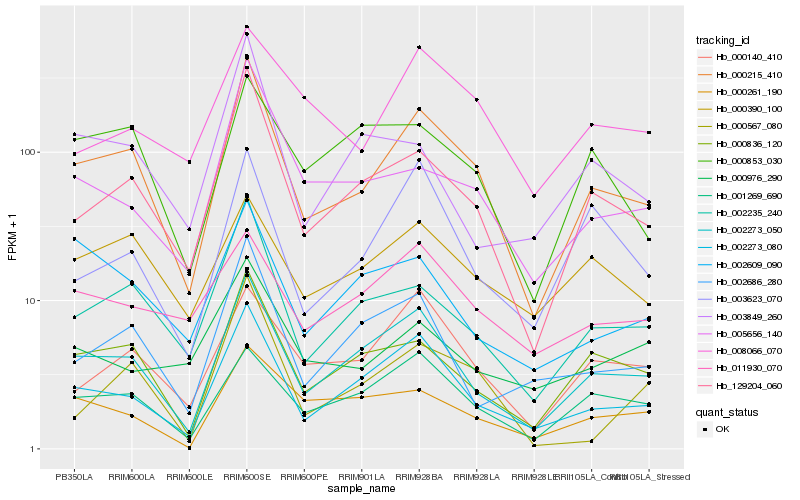
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_410 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 2 |
Hb_002273_050 |
0.1132030514 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_129204_060 |
0.1167512881 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 4 |
Hb_002273_080 |
0.1244593363 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 5 |
Hb_002235_240 |
0.1353019552 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 6 |
Hb_000836_120 |
0.1384527333 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 7 |
Hb_001269_690 |
0.167837291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005656_140 |
0.1711833557 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 9 |
Hb_000140_410 |
0.1773466657 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002686_280 |
0.1777257665 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_003623_070 |
0.1893864834 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_000567_080 |
0.1899061609 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 13 |
Hb_000390_100 |
0.1906652792 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 14 |
Hb_000261_190 |
0.19304809 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Jatropha curcas] |
| 15 |
Hb_000853_030 |
0.1938887135 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 16 |
Hb_008066_070 |
0.1965178181 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 17 |
Hb_002609_090 |
0.201416471 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_003849_260 |
0.2016692075 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 19 |
Hb_000976_290 |
0.2019133631 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 20 |
Hb_011930_070 |
0.202368353 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |