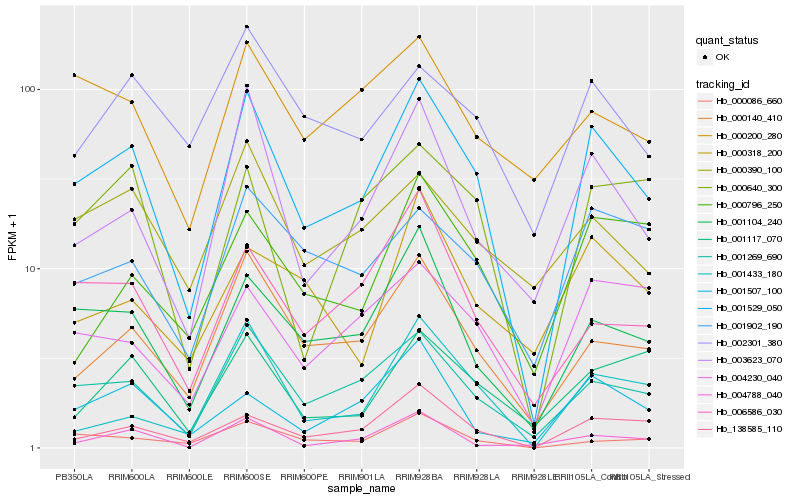
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001529_050 |
0.0 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 2 |
Hb_001269_690 |
0.123497946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001104_240 |
0.1484732434 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000140_410 |
0.1523932313 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 5 |
Hb_138585_110 |
0.1588313165 |
- |
- |
- |
| 6 |
Hb_003623_070 |
0.158842847 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_000086_660 |
0.1799203837 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 8 |
Hb_000640_300 |
0.1836795729 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 9 |
Hb_006586_030 |
0.1842119921 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 10 |
Hb_001902_190 |
0.1842359643 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 11 |
Hb_004230_040 |
0.1855955466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000390_100 |
0.1885146362 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 13 |
Hb_001507_100 |
0.1890965033 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_001433_180 |
0.1898587214 |
- |
- |
hypothetical protein JCGZ_23261 [Jatropha curcas] |
| 15 |
Hb_002301_380 |
0.190893981 |
- |
- |
PREDICTED: VQ motif-containing protein 4 [Jatropha curcas] |
| 16 |
Hb_001117_070 |
0.192243288 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 17 |
Hb_004788_040 |
0.1932324238 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000200_280 |
0.1969483995 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 19 |
Hb_000796_250 |
0.2000173635 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 20 |
Hb_000318_200 |
0.2022591888 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |