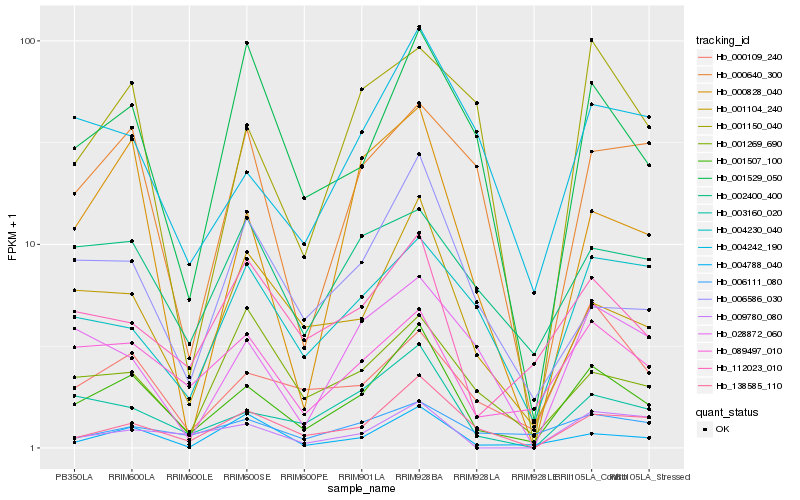
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001507_100 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_003160_020 |
0.153759729 |
- |
- |
PREDICTED: uncharacterized protein LOC105639457 [Jatropha curcas] |
| 3 |
Hb_001104_240 |
0.161730802 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_089497_010 |
0.1658322632 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_138585_110 |
0.1704424055 |
- |
- |
- |
| 6 |
Hb_004242_190 |
0.1840951432 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 7 |
Hb_112023_010 |
0.1873886437 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 8 |
Hb_001529_050 |
0.1890965033 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 9 |
Hb_006586_030 |
0.1944963863 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 10 |
Hb_000109_240 |
0.1952258805 |
- |
- |
PREDICTED: uncharacterized protein LOC102615432 [Citrus sinensis] |
| 11 |
Hb_004788_040 |
0.1971339469 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000828_040 |
0.200868492 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 13 |
Hb_002400_400 |
0.2052320071 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_006111_080 |
0.2064078177 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 15 |
Hb_028872_060 |
0.210381302 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 16 |
Hb_004230_040 |
0.2132023736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000640_300 |
0.2132447957 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 18 |
Hb_001150_040 |
0.2153034134 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Solanum lycopersicum] |
| 19 |
Hb_009780_080 |
0.2176033961 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 20 |
Hb_001269_690 |
0.2195331256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |