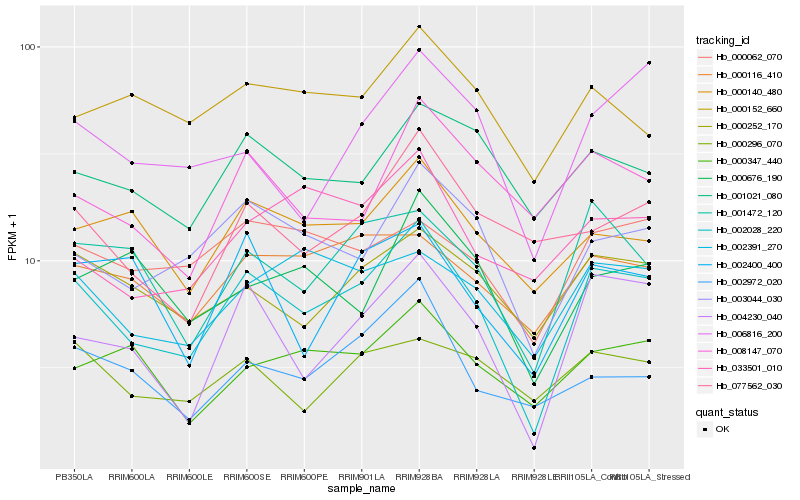
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002028_220 |
0.0 |
- |
- |
aminoadipic semialdehyde synthase, putative [Ricinus communis] |
| 2 |
Hb_004230_040 |
0.1207607208 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003044_030 |
0.1264479588 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Cicer arietinum] |
| 4 |
Hb_001472_120 |
0.141265795 |
- |
- |
- |
| 5 |
Hb_000252_170 |
0.1436569901 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002391_270 |
0.151308702 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 7 |
Hb_006816_200 |
0.1517744738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000062_070 |
0.153358066 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 9 |
Hb_001021_080 |
0.1534297166 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 10 |
Hb_000116_410 |
0.1552899406 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 11 |
Hb_000140_480 |
0.1560692695 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 12 |
Hb_000296_070 |
0.1581240626 |
- |
- |
hypothetical protein B456_003G048700 [Gossypium raimondii] |
| 13 |
Hb_008147_070 |
0.1584306378 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 14 |
Hb_077562_030 |
0.1590542494 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002400_400 |
0.1593196911 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_000347_440 |
0.1601753613 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 17 |
Hb_002972_020 |
0.1613214143 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 18 |
Hb_000152_660 |
0.1632226949 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 19 |
Hb_033501_010 |
0.1649437878 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 20 |
Hb_000676_190 |
0.1677954109 |
- |
- |
hypothetical protein JCGZ_08279 [Jatropha curcas] |