| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
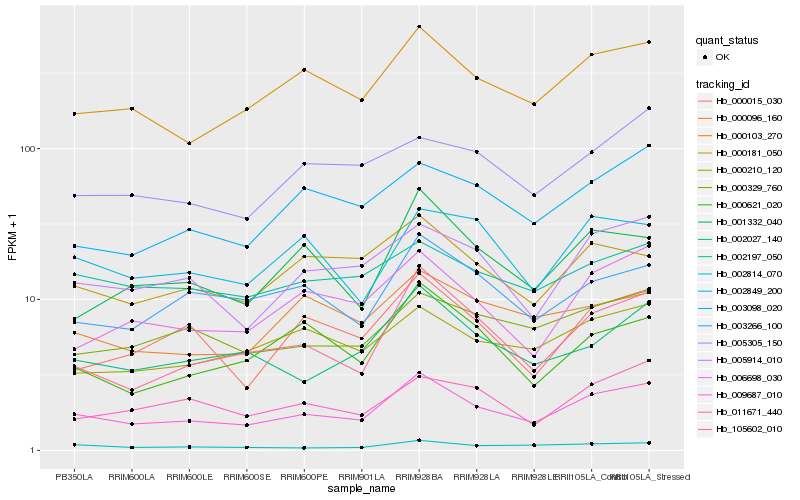
Hb_009687_010 |
0.0 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X4 [Jatropha curcas] |
| 2 |
Hb_000103_270 |
0.0828206066 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_005914_010 |
0.0957048189 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22 [Jatropha curcas] |
| 4 |
Hb_105602_010 |
0.0997602878 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 5 |
Hb_002027_140 |
0.1079587045 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |
| 6 |
Hb_006698_030 |
0.1080480876 |
- |
- |
PREDICTED: uncharacterized protein LOC105633969 [Jatropha curcas] |
| 7 |
Hb_000329_760 |
0.1097215415 |
- |
- |
PREDICTED: probable mitochondrial import inner membrane translocase subunit TIM21 [Jatropha curcas] |
| 8 |
Hb_003266_100 |
0.1097567026 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 9 |
Hb_000210_120 |
0.1118787532 |
- |
- |
PREDICTED: uncharacterized protein LOC105635829 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002814_070 |
0.1119560058 |
- |
- |
PREDICTED: uncharacterized protein LOC104877459 [Vitis vinifera] |
| 11 |
Hb_003098_020 |
0.1137504209 |
- |
- |
40S ribosomal protein S5B [Hevea brasiliensis] |
| 12 |
Hb_002197_050 |
0.1148037585 |
- |
- |
hypothetical protein CICLE_v10002157mg [Citrus clementina] |
| 13 |
Hb_000181_050 |
0.1168890529 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 14 |
Hb_000015_030 |
0.1173636552 |
- |
- |
PREDICTED: THO complex subunit 4B-like [Jatropha curcas] |
| 15 |
Hb_011671_440 |
0.117902696 |
- |
- |
ATP-dependent DNA helicase pcrA, putative [Ricinus communis] |
| 16 |
Hb_002849_200 |
0.1191166611 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 17 |
Hb_005305_150 |
0.1208600011 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 18 |
Hb_001332_040 |
0.1231024804 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 19 |
Hb_000096_160 |
0.1233017121 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 20 |
Hb_000621_020 |
0.123562809 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |