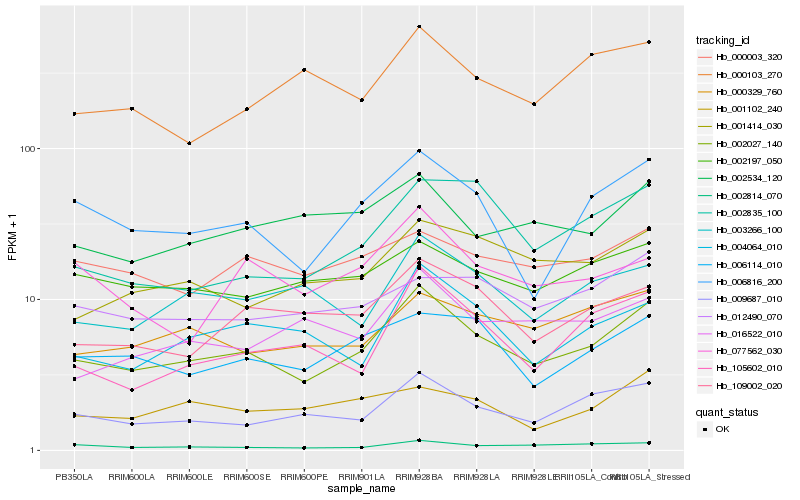
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002027_140 |
0.0 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |
| 2 |
Hb_009687_010 |
0.1079587045 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X4 [Jatropha curcas] |
| 3 |
Hb_006816_200 |
0.1101850587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_109002_020 |
0.1231790013 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 5 |
Hb_004064_010 |
0.1246204566 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |
| 6 |
Hb_006114_010 |
0.1264056867 |
- |
- |
PREDICTED: uncharacterized protein LOC105629068 [Jatropha curcas] |
| 7 |
Hb_003266_100 |
0.130601773 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 8 |
Hb_000329_760 |
0.1306170881 |
- |
- |
PREDICTED: probable mitochondrial import inner membrane translocase subunit TIM21 [Jatropha curcas] |
| 9 |
Hb_105602_010 |
0.1309136392 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 10 |
Hb_002814_070 |
0.1309237651 |
- |
- |
PREDICTED: uncharacterized protein LOC104877459 [Vitis vinifera] |
| 11 |
Hb_001414_030 |
0.1339259405 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 12 |
Hb_002534_120 |
0.135321845 |
- |
- |
PREDICTED: protein DCL, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002197_050 |
0.1378872014 |
- |
- |
hypothetical protein CICLE_v10002157mg [Citrus clementina] |
| 14 |
Hb_000003_320 |
0.1409578374 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 24 [Jatropha curcas] |
| 15 |
Hb_012490_070 |
0.142172705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_016522_010 |
0.1430210648 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 17 |
Hb_077562_030 |
0.143158445 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002835_100 |
0.143180133 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 19 |
Hb_001102_240 |
0.1459733037 |
- |
- |
PREDICTED: NADH kinase isoform X1 [Jatropha curcas] |
| 20 |
Hb_000103_270 |
0.1461673069 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |